Early detection and diagnosis of cancer is essential to give sufferers an increased chance of overcoming the disease. The symptoms of liver cancer, in particular, are fairly innocuous – tumours are difficult to detect by physical examination and therefore the disease is not usually discovered until the later stages of development.
Dong Wang and co-workers have developed an electrochemical immunoassay which has the potential to improve the early detection rates of liver cancer by simultaneous detection of not one, not two, but three tumour markers. This simultaneous multianalyte immunoassay (SMIA) has a number of advantages over single-analyte immunoassay methods such as reduced overall cost per assay; improved efficiency; the potential to quantitatively measure the concentrations of proteins detected; as well as having a panel of biomarkers to confirm the diagnosis, lowering the likelihood of false-positive or false-negative results.
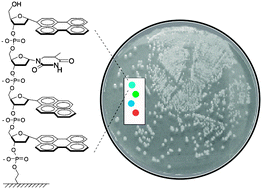
The team selected three electrochemical redox species with distinct voltammetric peaks to label three different antibodies as signal tags. These were then loaded onto carbon nanotubes coated with gold nanoparticles to improve the signal response.
Binding of the redox species labelled nanotube-antibody conjugates to the cancer biomarkers enables their electrochemical detection
By deciphering the voltammetric read-out, the team were able to establish successful binding events between the probes and the protein biomarkers, allowing simultaneous identification of the three analytes. Wang and his team then increased the sophistication of the immunoassay further by proving quantitative detection.
Dong Wang and co-workers appeared to have produced a robust SMIA with low detection limits that has the potential to save many lives. This efficient, gold nanoparticle-based technology could also be applied to other types of cancer or diseases – an immunoassay with a Midas touch.
To find out more, download the ChemComm article.
Posted on behalf of Sarah Brown, ChemComm web science writer.










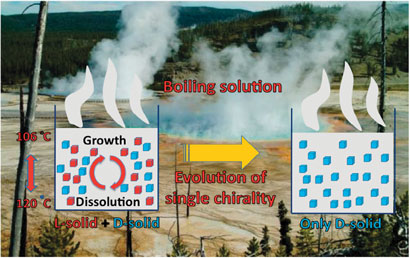 The boiling solutions in prebiotic hot springs could shed light on the emergence of a single chiral form of biomolecules in nature, say Spanish scientists.
The boiling solutions in prebiotic hot springs could shed light on the emergence of a single chiral form of biomolecules in nature, say Spanish scientists.