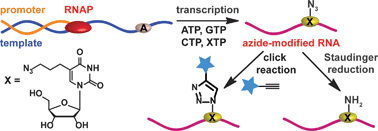
If you can’t beat them, join them… or rather incorporate, and then join them together. This is the approach adopted by Srivatsan and colleagues for conjugating labels to RNA, which they achieve by incorporating azide functionality into RNA nucleotides.
DNA oligonucleotides are routinely labelled by incorporating modified nucleosides into the desired DNA sequence and introducing the label post-synthesis; however, these standard methods offer low yields of the desired conjugate. Furthermore, modification of RNA is more challenging than DNA modification due to its inherent instability.
Copper catalysed alkyne–azide cycloadditions (CuAAC) and Staudinger ligations are the conjugation reactions du jour due to their high chemoselectivity and reported high yield. So, with this in mind, were Srivatsan and his team able to incorporate the required moieties into RNA oligonucleotides?
Yes! Using some extremely nice chemistry, they synthesised an azide-modified nucleotide and incorporated it into an oligoribonucleotide using in vivo transcription reactions in the presence of a series of promoter-template oligonucleotides. After rigorous testing and analysis of their modified nucleic acid sequence, click reactions were performed to yield biotinylated and fluorescent-labelled click products. They also showed that amine functionality can be introduced through Staudinger ligation.
Although azides are not usually found in nature, they are becoming more useful and versatile in the design of diagnostic and therapeutic biological probes as has been very elegantly demonstrated here.
Read more in Srivatsan’s ChemComm article…
Also of interest: Synthetic DNA synthesises RNA by transcription: ‘click’ here for more
Posted on behalf of Sarah Brown, web science writer for ChemComm.