A new trend gripping popular science (and business) is personal DNA testing. A host of companies sell the idea that your genetic heritage distinguishes you, and reveals who you really are. Do you really want to know what you are on a molecular level? If it’s a numbers game, you are mostly bacterial (that’s why you’re so cultured!). Bacteria outnumber our body’s cells 1.3:1. The majority of bacteria reside in the gastrointestinal tract, contributing to a microbiota of archea, eukaryotes and viruses that have a commensal relationship with the human body.
The microbiota has been linked to many vital physiological pathways including human nutrition (harvesting further energy and nutrients), immunity, inflammation and the detoxification of xenobiotic substances. Conversely, dysregulation of such pathways has been associated with diseases such as diabetes, cancer, inflammatory bowel syndrome and cardiovascular disease. Due to the number of pathways involved, it is hoped that studying the microbiota may identify enzyme targets for therapeutics or biomarkers for disease.
However, understanding of these pathways is limited, and research progress relies on advances in analytical tools. Headed by Dr Daniel Globisch at Uppsala University in Sweden, researchers have developed an analytical method to identify O-sulphated metabolites. Microbes are capable of a suite of metabolic reactions that complement the capabilities of human enzymes, and O-sulphate functionalization is characteristic of microbe/human co-metabolism as it can be catalysed by bacterial sulphotransferase enzymes.
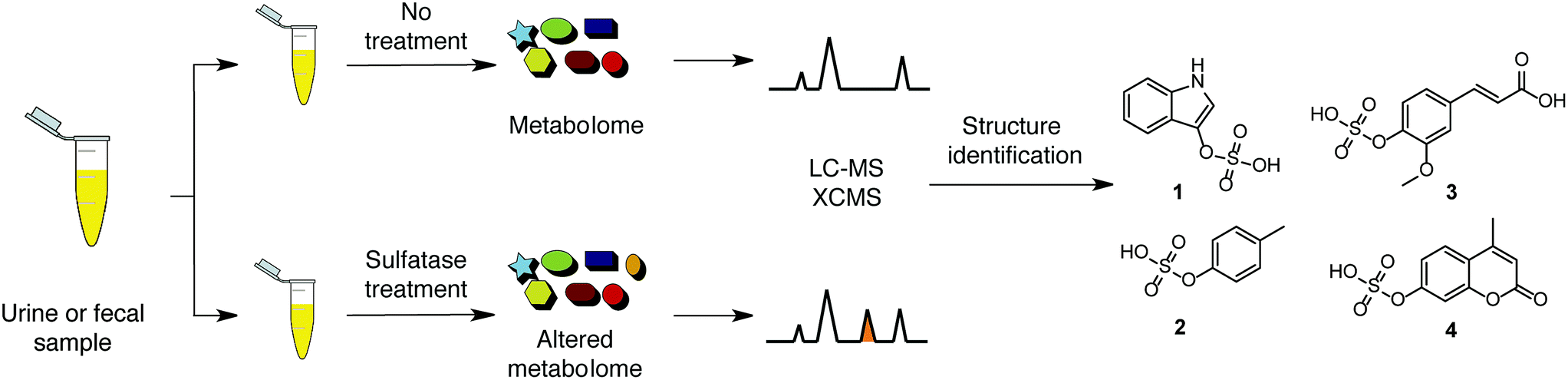
O-sulphate functionalised molecules can be selectively analysed using sulphatase treatment and UPLC-MS/MS
The researchers developed an assay to identify O-sulphated compounds in urine and faecal samples. The assay was designed using a sulphatase enzyme capable of hydrolysing the oxygen-sulfur bond in a wide variety of arylsulphate molecules. Samples were treated with this enzyme and the resulting mixtures were analysed by UPLC-MS/MS (ultra performance liquid chromatography/tandem mass spectrometry). Results were compared with the data output of control samples without enzymatic treatment and features in the spectra with a mass change of 79.9568 m/z (loss of the sulfate group) were identified, leading to the identification of 206 O-sulphated metabolites. This is a notable result as it triples the number of sulphated metabolites currently recorded in the human metabolome database.
A number of interesting compounds were identified: ferulic acid is a metabolite produced by the microbiota thought to prevent thrombosis and artherosclerosis, and indoxyl sulfate and p-cresylsulfate are biomarkers of chronic kidney disease and cardiovascular disease, respectively. Mentioned are three molecules of the 206 uncovered, giving a glimpse of the applications that further research, armed with the right analytical tools, might discover.
To find out more please read:
Caroline Ballet, Mário S. P. Correia, Louis P. Conway, Theresa L. Locher, Laura C. Lehmann, Neeraj Garg, Miroslav Vujasinović, Sebastian Deindl, J.-Matthias Löhr, Daniel Globisch.
Chem. Sci., 2018, Advance Article
DOI: 10.1039/c8sc01502c

About the author
Zoë Hearne is a PhD candidate in chemistry at McGill University in Montréal, Canada, under the supervision of Professor Chao-Jun Li. She hails from Canberra, Australia, where she completed her undergraduate degree. Her current research focuses on transition metal catalysis to effect novel transformations, and out of the lab she is an enthusiastic chemistry tutor and science communicator.