In this new Perspective, Chemical Science Associate Editor Tom Muir of Princeton University elucidates the biological role and evolutionary origin of inteins – a fascinating class of proteins which have the ability to “process”, or sever, their own peptide backbone. Usually, this type of post-translational modification is done enzymatically by proteases, but inteins contain a module which allows the spontaneous scission of peptide bonds with no external factor or energy source required. The only things needed are certain chemical functional groups in the neighbouring peptide residues, and the correct spatial folding of the domain.
-
Obviously, this is a truly interesting process, as the functions and actions of a protein are determined by its amino acid sequence and structure. By breaking peptide bonds and creating new ones, inteins essentially act as an on/off switch for the protein, the potential application for which is staggering.
The intein is flanked on either side by two exteins; during the “processing” reaction, the bonds between intein and exteins are broken, and a new bond between the exteins is created. Inteins share a common biochemical mechanism for this process, which is illustrated below. Crucially, all inteins must contain a cysteine or serine at their N-terminus which provides the nucleophile for the initial acyl shift. A subsequent trans(thio)esterification and an additional acyl shift forms the spliced product and the excised intein.
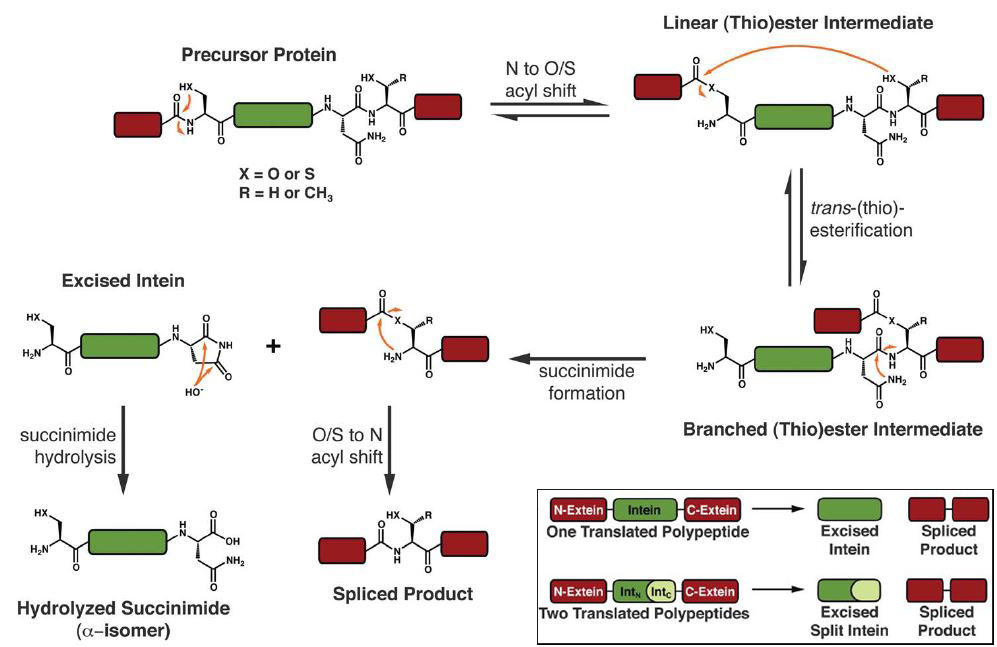
- Mechanism of protein splicing
Muir and co-author Neel Shah investigate the evolutionary origin of inteins, which can be found in all domains of life – eukaryotes, bacteria, archaea and viruses. Although they are often found in proteins involved in genetic “housekeeping” – DNA replication, transcription, and maintenance – inteins have no obvious biological role, and do not provide any benefit to the host organism. As such, they are known as “selfish” genes. Despite the mystery of their purpose and origin, what is clear is that the future is bright for inteins.
-
The authors discuss a number of applications of inteins, the most exciting of which is conditional protein splicing (CPS) where inteins can be used as an on/off switch for the proteins they are splicing, even in vivo. CPS is currently achieved in a number of ways, shown below, in which the intein is kept in an inactive state – via (a) conformational distortion, (b) caging of the active site, or (c) the physical separation of a split intein – until activation is desired. Ligand binding, deprotection or dimerisation, respectively, then releases the active intein and triggers the peptide splicing. CPS is a promising tool for cell biology and should facilitate the development of “smart” protein therapeutics that are activated only at the target site.
All in all, this Perspective makes for an interesting read on a class of proteins with impressive potential. I think it’s a safe bet to predict that when it comes to future smart therapeutics, inteins will definitely make the cut.
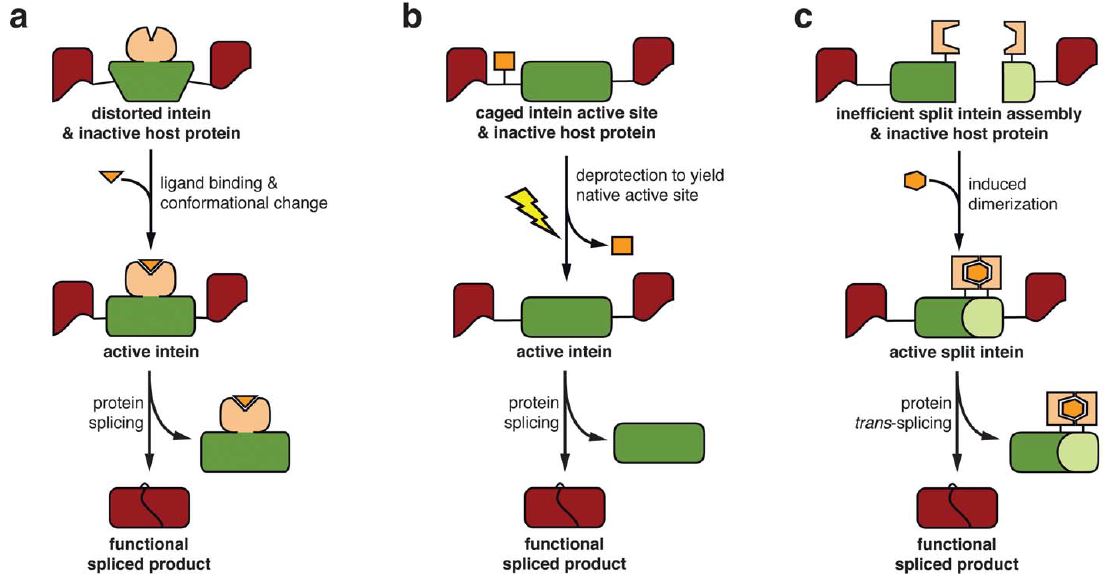
- Conditional protein splicing (CPS): a) Allosteric intein activation by ligand binding; b) Intein activation via deprotection of a photo-caged active site residue; c) Activation via chemically-induced dimerisation
For more, you can read Muir and Shah’s Chemical Science Perspective here:
Professor Muir serves as one of Chemical Science‘s Associate Editors, handling submission in chemical biology – read more about him and what useful advice he wishes someone had told him as an undergraduate.
Our Associate Editors Tom Muir and Ben Davis have highlighted their recommended chemical biology papers on Chemical Science – read their Editor’s Choice selection for FREE today!
Find many more excellent articles on chemical biology here: Online collection: Chemical biology
Ruth E. Gilligan is a guest web-writer for Chemical Science. She recently completed her PhD in the group of Prof. Matthew J. Gaunt at the University of Cambridge, and is currently pursuing an internship at Science Foundation Ireland.