Hydrogen bonding is all around us. The intermolecular force of attraction between a hydrogen atom bound to an electronegative centre (the hydrogen bond donor, HBD) and another nearby electronegative atom with a lone pair of electrons (the hydrogen bond acceptor, HBA) is present in many chemical structures and can be seen in many biological motifs such as in enzymes or proteins. Beyond a simple HBD-HBA pair (Figure 1a), hydrogen bonding can cascade between multiple HBD-HBA pairs (Figure 1b). In these paired units, the presence of a central proton mediator (e.g. imidazole) can induce polarisation to make the resulting hydrogen bonds stronger, promoting further reactivity and selectivity (Figure 1c).
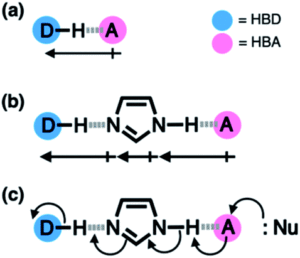
Figure 1. Hydrogen bonding between donor (D) and acceptor (A) atoms, with the net dipole shown by the arrows beneath. (a) Simple HBD-HBA pair. (b) Cascade hydrogen bonding around a central imidazole proton mediator, that can promote further reactivity with a larger net additive dipole (c).
Some enzymes cleverly make use of cascade hydrogen bonding to control the strength of the hydrogen bonds that form between the limited number of available amino acids. One example is a class of enzymes with a ‘catalytic triad’, whereby a hydrogen bonding array exists between the hydroxyl group of serine, the imidazole group of histidine and the carboxylate group of aspartate residues (Figure 2a). Researchers from South Korea took inspiration from such catalytic triads to create a ‘synthetic triad’ with a biomimetic hydrogen bonding network (Figure 2b, compound 1). The researchers employed a central benzimidazole to their synthetic triad to act as a platform to align the HBD-HBA pairs, instead of the precise three-dimensional structure that would anchor these pairs in enzymes.
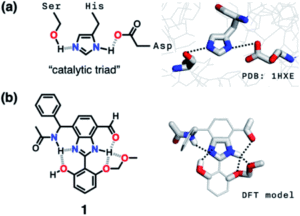
Figure 2. (a) Chemical (left) and X-ray (right) structures of the ‘catalytic triad’ in the active site of the enzyme serine protease. (b) Chemical structure (left) and computational model (right) of the ‘synthetic triad’ designed by the researchers.
The researchers envisioned that their biomimetic small molecule could be used as a fluorescent probe owing to the photophysical properties of the chosen benzimidazole motif. They designed the probe for cyanide detection, where capture of a toxic cyanide ion turns on fluorescence in the probe (Figure 3c). The design of the probe was therefore influenced with the target application in mind, so the researchers systematically added each HBD-HBA pair around the benzimidazole, as shown in Figure 3b.
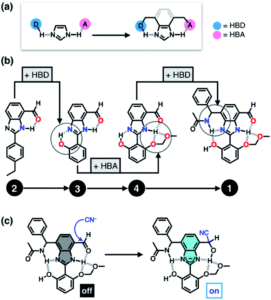
Figure 3. (a) Cascade hydrogen bonding around a benzimidazole core. (b) The systematic design of the probe, starting from one HBD-HBA pair up to four pairs. (c) Mechanism of capture of the cyanide ion to turn on fluorescence.
An aldehyde functional group was selected for the first HBD-HBA pair, due to its ability to form hydrogen bonds that can quench the fluorescence in the absence of cyanide (compound 2). The researchers tested compound 2 for cyanide detection and indeed observed fluorescence, but found that the fluorescence also occurred in the presence of a simple Brønsted base. The design of the probe was then iteratively modified until fluorescence was selective for cyanide addition, with a total of four HBD-HBA pairs around the benzimidazole centre that mutually reinforced one another. This strategy shows promise for the design of other fluorescent probes and could also be utilised for other biological targeting applications.
To find out more, please read:
Biomimetic hydrogen-bonding cascade for chemical activation: telling a nucleophile from a base
Hyunchang Park and Dongwhan Lee*
Chem. Sci., 2021, Advance Article
About the blogger:
 Dr. Samantha Apps recently finished her post as a Postdoctoral Research Associate in the Lu Lab at the University of Minnesota, USA, and obtained her PhD in 2019 from Imperial College London, UK. She has spent the last few years, both in her PhD and postdoc, researching synthetic nitrogen fixation and transition metal complexes that can activate and functionalise dinitrogen. Outside of the lab, you’ll likely find her baking at home, where her years of synthetic lab training has sparked a passion in kitchen chemistry too.
Dr. Samantha Apps recently finished her post as a Postdoctoral Research Associate in the Lu Lab at the University of Minnesota, USA, and obtained her PhD in 2019 from Imperial College London, UK. She has spent the last few years, both in her PhD and postdoc, researching synthetic nitrogen fixation and transition metal complexes that can activate and functionalise dinitrogen. Outside of the lab, you’ll likely find her baking at home, where her years of synthetic lab training has sparked a passion in kitchen chemistry too.