Have you ever heard of nonulosonic acids? As a materials chemist I hadn’t, as most of the acids I’ve dealt with are either acting as ligands or inorganic and I tend to forget that sugars are actually acids. These nine-carbon sugars are critical for a wide range of cellular functions throughout living organisms. Over 100 different members of this broad group of acids have been identified, all of which can undergo diversification at multiple positions, generating an even larger library of derivatives. What types of modifications can occur has yet to be fully realized, as novel compounds are continually discovered, particularly in prokaryotes. Even when examining a limited number of modifications, say 15, the number of possible derivatives is staggering, reaching several thousand without stereochemical considerations. Identifying these acids is key, as they have been linked to virulence in pathogenic bacteria and their previously mentioned diversity makes them challenging to genomically analyze.
Given these challenges, current detection practices rely on staining and fluorescent labeling which can be labor intensive and specific to a small number of acids. Early approaches based on mass spectroscopy were limited due to the complex backgrounds produced and the difficulties separating signal and noise. However, developments in high-resolution spectrometers and increased data processing abilities have led to successful analysis of increasing numbers of known metabolites. Researchers in the Netherlands and Russia extended this work to develop a screening approach to identify a diverse range of nonulosonic acids via small mass channel mass spectroscopy from previously unexplored environmental microbes (Figure 1).
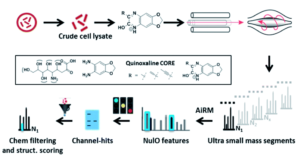
Figure 1. Outline of the general procedure for the survey pipeline, beginning from the cells to the final structural analysis of the identified acids.
To successfully identify these unknown derivatives, the researchers identified specific ulosonic acid fragments invariant to functionalization. They used 1,2-diamino-4,5-methylene dioxybenzene, an alpha-keto-acid specific labeling reagent, to shift the double bond equivalents from the general cellular background and help conserve the core ulosonic acid structure. By screening a known range of acids with different functional group moieties, they were able to identify universal features for this category of molecules (Figure 2). From there, they created a full process for rapid-throughput studies that automated everything from initial identification of a potential acid molecule through filtering and structural analysis.
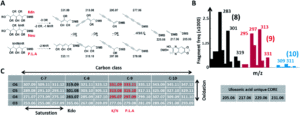
Figure 2. A) Universal fragmentation route used to identify nonulasonic acids in the screening, B) data output of various ulosonic acid derivatives identified in the study, and C) differences seen from the chemical labeling introduced in the procedure.
After the pipeline was established, the researchers used various standard cell lines, plant, animal, and algal, for a molecular level survey to confirm the validity of their approach for full-cell analysis. In general, the plants and yeast cells contained no nonulosonic acids, as expected, while the microalgae and animal cells had a range of different nonulosonic acids present. As these acids are thought to play a role in bacterial virulence, a bacteria known to display the acids was screened and several subtypes of animal and bacterial acids were identified as incorporated into the cells. With that validation, the researchers moved on to a wide range of non-pathogenic bacteria. They discovered that over half of these bacteria possess nonulosonic acids; in fact, some have only slightly lower acid abundance than mammalian cells. While much of this data was obtained from single cultures, it represents exciting validation of a broad new approach that could be used to identify potential targets for medical applications and continue to extend our understanding of the diversity of nonulosonic acids.
To find out more, please read:
Hugo B. C. Kleikamp, Yue Mei Lin, Duncan G. G. McMillan, Jeanine S. Geelhoed, Suzanne N. H. Naus-Wiezer, Peter van Baarlen, Chinmoy Saha, Rogier Louwen, Dimitry Y. Sorokin, Mark C. M. van Loosdrecht and Martin Pabst
Chem. Sci., 2020, 11, 3074-3080.
About the blogger:
 Dr. Beth Mundy is a recent PhD in chemistry from the Cossairt lab at the University of Washington in Seattle, Washington. Her research focused on developing new and better ways to synthesize nanomaterials for energy applications. She is often spotted knitting in seminars or with her nose in a good book. You can find her on Twitter at @BethMundySci.
Dr. Beth Mundy is a recent PhD in chemistry from the Cossairt lab at the University of Washington in Seattle, Washington. Her research focused on developing new and better ways to synthesize nanomaterials for energy applications. She is often spotted knitting in seminars or with her nose in a good book. You can find her on Twitter at @BethMundySci.