The idea of using DNA-based devices to create highly specific sensors, diagnostic tools, and therapeutics has inspired widespread research. DNA molecular switches, a basic class of DNA nanostructures, turn some process on and off depending on whether a substrate binds to the switch. They are typically coupled with some form of signal amplification process to increase sensitivity and should theoretically provide enhanced signal-to-noise. Unfortunately, current amplification procedures have a significant amount of background reactions, called leakage, that limit their current utility. One approach to design better switches is to find ways to observe single molecule dynamics. Combining transient binding of complimentary oligonucleotides with high resolution fluorescence microscopy allows for the development of highly sensitive molecular switches without leakage problems.
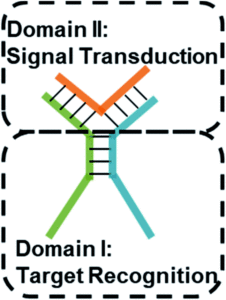
To do this, researchers in China developed a series of three-way junction DNA-based molecular switches. These TWJs possess a recognition domain, which interacts with the target of interest and induces a structural change in the TWJ, and a transduction domain, which then becomes accessible and binds to a fluorescent molecule (Figure 1).
Experimentally, the TWJs were captured on the imaging surface and only fluorophores bound to a TWJ would remain in place long enough for signal to be acquired by the camera with a 500 ms integration time. The researchers found that shorter transduction domains with 5 or fewer base pairs were not stable enough to allow the fluorescent probes access in the absence of a bound target. The next generation of TWJs feature a hybridization probe to allow the switch to recognize specific DNA inputs. In the presence of inputs, the researchers observed transient binding behavior of the fluorophore, whereas they observed only nonspecific binding in the absence of inputs. The ability to differentiate between non-specific and transient binding in the single-molecule system gives a detection limit of 10 fM without concerns about leakage.
Building on this work, the researchers utilized the same general framework and substituted aptamer sequences for the hybridization probe in the recognition domain. They utilized split aptamer fragments that only draw together when bound to a target molecule. This motif was tested on ATP, a small molecule, and thrombin, a protein. These aptamer-coupled TWJs exhibited sensitivity to concentrations as low as 20 – 50 pm with high sensitivity (Figure 2). In the presence of molecular analogs to ATP or thrombin, the signal level showed no significant difference from that of a blank.
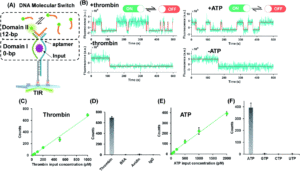
Figure 2. A) Split aptamer-based molecular switch schematic. B) Single-molecule fluorescence-time trajectory data in the presence (top) or absence (bottom) of targets. C) and D) Linear relationship between thrombin concentration and signal and specificity when compared to analogs. E) and F) Linear relationship between ATP concentration and signal and specificity when compared to analogs.
Another advantage of this system is its ability to provide information on the binding affinity of substrates, as it should impact the kinetics of the fluorescent probes. The dwell times of the fluorescence on and off states demonstrated exponential trends with changing input concentrations and could be fit to extract time constants. These time constants can then be used to derive the kinetics parameters and binding affinities of the target species. The general stability of the molecular switch framework allows for studying these types of interactions in a range of pH and salinity conditions, useful for mimicking different environments relevant to future applications. This provides a platform for studying the fundamental interactions that will allow DNA-based nanotechnology to move forward.
To find out more, please read:
Single-molecule dynamic DNA junctions for engineering robust molecular switches
Shuang Cai, Yingnan Deng, Shengnan Fu, Junjie Li, Changyuan Yu and Xin Su
Chem. Sci., 2019, 10, 9922-9927.
About the blogger:
 Beth Mundy is a PhD candidate in chemistry in the Cossairt lab at the University of Washington in Seattle, Washington. Her research focuses on developing new and better ways to synthesize nanomaterials for energy applications. She is often spotted knitting in seminars or with her nose in a good book. You can find her on Twitter at @BethMundySci.
Beth Mundy is a PhD candidate in chemistry in the Cossairt lab at the University of Washington in Seattle, Washington. Her research focuses on developing new and better ways to synthesize nanomaterials for energy applications. She is often spotted knitting in seminars or with her nose in a good book. You can find her on Twitter at @BethMundySci.