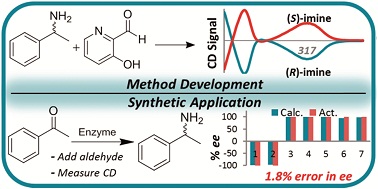
Aristotle’s saying “the whole is more than the sum of its parts” is often well-exemplified in supramolecular chemistry – the result of non-covalent interactions present in a complex assembly of simple building blocks often generate novel properties that would not otherwise be present in a simple mixture of the components. The design of systems that exhibit these emergent phenomena is therefore an aim of many supramolecular chemists, including researchers from India who have achieved the control of supramolecular helicity in a bistable artificial self-assembled system through the addition of either a chiral or achiral component. This example of allosteric regulation in a synthetic system is an interesting analogue of that which occurs in biological systems, and offers an attractive approach to inducing not only conformational change in a pre-formed assembly, but also new chemical properties that could not otherwise be observed.
In biological systems, allosteric regulation, the binding of a secondary molecule to an enzyme, is employed to modulate the conformation of the active site and thus the effectiveness of an enzyme. This feature allows for communication between remote sites in a cell by providing a control loop in enzymes. It is therefore of interest to scientists to apply this allosteric regulation synthetically as a way of introducing a dynamic component to a molecular system.
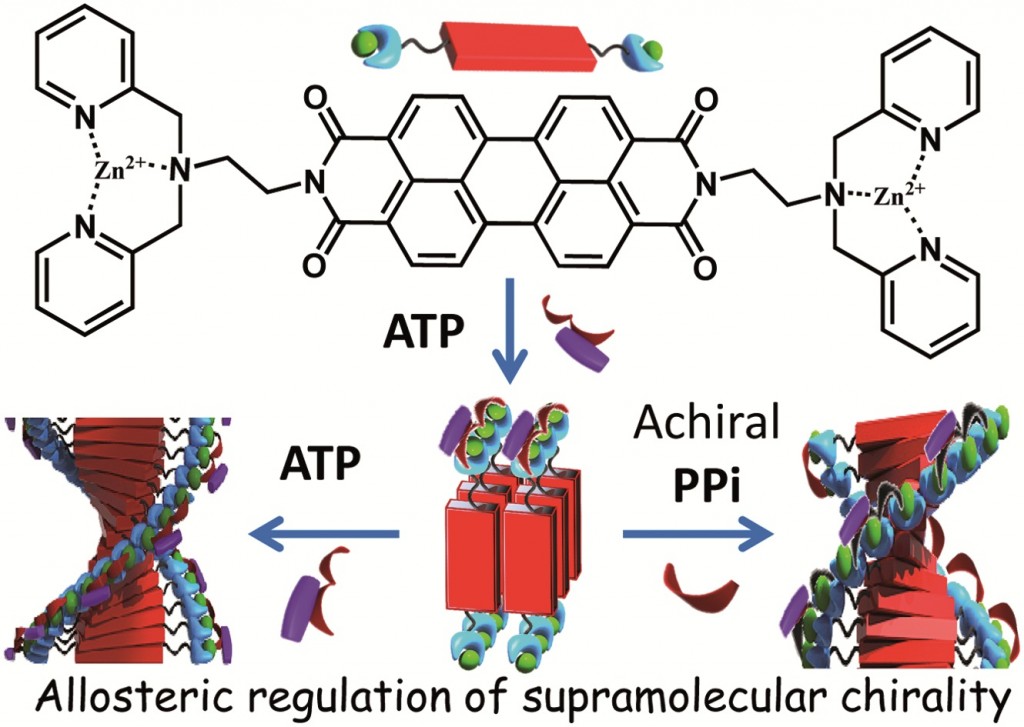
Mohit Kumar and Subi George at the Jawaharlal Nehru Centre for Advanced Scientific Research[EB1] (JNCASR) in India have begun to experiment with the use of allostery in synthetic system[EB2] in order to induce [EB3] helicity in a polymeric supramolecular assembly comprised of a perylene bisimide functionalised at either end by a Zinc(II) motif that is known to bind phosphate ligands. In the “off” state this assembly exists as linear stacks of the molecule, such that overall the assembly is achiral. Upon the binding of 0.5 equivalents of chiral adenine trisphospate (ATP), a supramolecular reorganization is observed to form the helically dormant H1 state, which, upon the addition of further equivalents of ATP, results in a second conformational change to the helical H2, whereby the linear stacks are now twisted in nature and exhibit chirality. This example of chiral induction, in which a single molecule can turn “on” its chirality, exemplifies homotropic allosteric regulation. Furthermore, upon the addition of a second achiral pyrophosphate ligand (PPi) to H1, a different helical H2 state is induced in the assembly. The introduction of two chemically different molecules that interact with the assembly in different ways is an example of heterotropic allosteric regulation.
This use of supramolecular chemistry to post-synthetically elicit a change in a molecular assembly is an exciting way of mimicking the allostery observed in biology. Even more remarkable, is the ability of one system to respond in two different ways.
Read this HOT ChemSci article in full for free*!
Homotropic and heterotropic allosteric regulation of supramolecular chirality
Mohit Kumar and Subi B. George
DOI: 10.1039/C4SC00813H
About the Writer
Anthea Blackburn is a guest web writer for Chemical Science. Anthea is a graduate student hailing from New Zealand, studying at Northwestern University in the US under the tutelage of Prof. Fraser Stoddart (a Scot), where she is exploiting supramolecular chemistry to develop multidimensional systems and study the emergent properties that arise in these superstructures. When time and money allow, she is ambitiously attempting to visit all 50 US states before graduation.
*Access is free untill 06.06.14 through a registered RSC account – click here to register