At the mention of DNA most people think of the storage of genetic information, but due to its biocompatibility and the ease with which it can be manipulated, it is becoming increasingly common for DNA to be used as a building material in nanotechnology.
DNA cages have attracted interest in the world of drug delivery as they can encapsulate small molecule drugs and are easily taken up by cells. The precision with which the size and shape of the cages can be controlled is another attractive aspect. Past studies have established that DNA cages can target cells, deliver encapsulated cargo and have a cytotoxic effect in cancer cells. However, specific control of the cellular delivery profile of DNA cages has not yet been achieved.
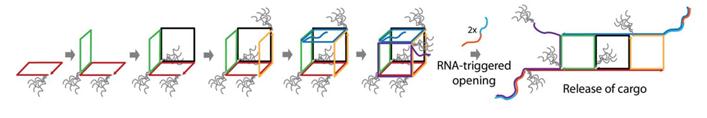
Sleinman and co-workers, from McGill University based in Montréal, Canada, have created dynamic DNA cubes which ‘unzip’ in a specific cellular environment. The cages are assembled from six DNA strands which make up the six sides of the cube and only disassemble in the presence of a trigger found in prostate cancer cells. The authors tested the uptake and disassembly, or ‘unzipping’, of their DNA cubes in vitro using three different mammalian cell lines.
Hydrophobic and hydrophilic dendritic chains were added to the cube after initial testing. It was determined that these chains coat the exterior of the cube and have a significant effect on the uptake of the structure. While the addition of hydrophobic chains to the cube increase uptake, addition of hydrophilic chains increase the stability of the cube in cellular environments.
One of the exciting aspects about this work is the scope for adaptation; the cubes could potentially be designed to respond to any nucleic acid sequence found specifically in diseased cells. Future work in the group will also look at encapsulating and delivering cargo, such as oligonucleotide drugs, using this new delivery system.
To download the full article for free* click the link below:
Sequence-responsive unzipping DNA cubes with tunable cellular uptake profiles
Katherine E. Bujold, Johans Fakhoury, Thomas G. W. Edwardson, Karina M. M. Carneiro, Joel Neves Briard, Antoine G. Godin, Lilian Amrein, Graham D. Hamblin, Lawrence C. Panasci, Paul W. Wiseman and Hanadi F. Sleiman
DOI: 10.1039/C4SC00646A
*Access is free until 02/06/2014 through a registered RSC account – click here to register