If you thought squid only belonged in the depths of the ocean, think again. SQUID, or superconducting quantum interference device, has been used to study single-molecule magnets (SMMs) in solution, which could help us store more information on hard drives in future.
As the demand for increased information storage continues to rise, scientists have turned to the development of new nanostructured materials, incorporating SMMs. These tiny magnets can store information depending on the charge and spin properties of their electrons. But until now, there have been few studies to examine how much of these molecules’ magnetic properties come from their molecular properties and how much comes from the way they are packed together in the solid-state.
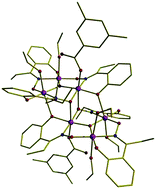
Using SQUID, which is a very sensitive device for detecting weak magnetic fields, Euan Brechin and colleagues have studied the spin properties of frozen solutions of two different hexanuclear manganese (Mn6) complexes. The two compounds display different spin-relaxation properties in the solid-state, but similar spin-dynamics once in solution. Brechin believes the study demonstrates that the SMM behaviour is intrinsically a molecular effect that can be modulated in the solid-state by crystal packing strain effects.
Read the full story in the Chemical Science Edge article, which, like all Chemical Science articles, is free to download until the end of 2011.
Be seen with the best – submit to Chemical Science today.