40 years after it was first isolated, UK scientists have identified the biosynthetic genes of an important antibiotic, offering insights into its poorly understood biosynthetic pathway.
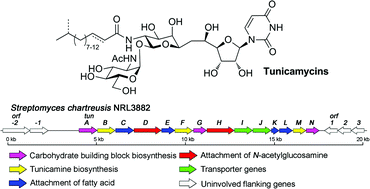
Tunicamycin antibiotics have attracted much attention (over 8000 citations) due to their unusual structure and potent inhibition of bacterial cell wall biosynthesis. Although they have been chemically synthesised, the lack of a sequence for the tunicamycin gene cluster (or any part of it) has left scientists puzzled over its biosynthetic pathway.
Now Benjamin Davis, at the University of Oxford, and colleagues have identified the tunicamycin biosynthesis genes in Streptomyces chartreusis, a soil bacterium, using genome sequencing and mining. Using this genetic insight, they have proposed the detailed biosynthetic pathway to this family of antibiotics.
The studies unlock a comprehensive and unusual toolbox of biosynthetic machinery with which to create variants of this natural product, says Davis. He anticipates this will lead to future therapeutic antibiotics with improved antibacterial activity and reduced cytotoxicity.
Find out more in Davis’ Chemical Science Edge article, downloadable for free. Access our free content any time, any place by registering for an RSC Publishing personal account today.