We are very pleased to introduce Rajendra Joshi and the team of authors of the paper ‘Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2‘. Their article has been very well received and handpicked by our reviewers and handling editors as one of our HOT articles. The team told us more about the work that went into this article and what they hope to achieve in the future. You can find out more about the authors and their article below and find more HOT articles in our online collection.

About Dr Rajendra Joshi
Dr. Rajendra Joshi received his Ph.D. in Biochemistry from National Chemical Laboratory, Pune, India in 1994. He has been associated with the area of Biotechnology & Bioinformatics for the about 28 years. He is presently serving as a Senior Director and Head of the Department, High Performance Computing-Medical and Bioinformatics Applications Group, at C-DAC, Pune.
His major area of expertise, is in the use of high performance parallel computers for biological research. His unique strength is in the form of good knowledge of biology and parallel computing. His main research interests include, molecular dynamics simulations of nucleic acids & proteins, genome sequence analysis, metabolic pathways and development of Problem Solving Environments. He has around 66 publications in internationally peer reviewed journals.
Could you briefly explain the focus of your article to the non-specialist (in one or two sentences only) and why it is of current interest?
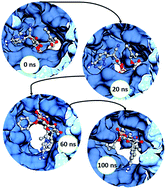
Remdesivir, the emergency drug approved for treating COVID-19 patients helps in blocking the multiplication of SARS-CoV-2 virus. The action of this drug on the viral protein RNA dependent RNA polymerase was mimicked using computational methods, namely, molecular docking and molecular dynamics simulations.
How big an impact could your results potentially have?
The predicted mechanism of action of the drug, remdesivir, on the viral protein of SARS-CoV-2 would help in designing inhibitor molecules against the viruses. The drug target protein is the one, which is observed to be the most conserved among the coronavirus family. Statistically significant results produced through computational drug repurposing methods, add to the prediction accuracy of the drug-target interactions that are of major interest to develop therapeutics.
Could you explain the motivation behind this study?
The need to find the best solution against the global pandemic was the biggest motivation behind this study. This work was performed in the month of May 2020 and during that time, remdesivir, was being considered as one of the best solutions to treat the COVID-19 patients until the designing of the vaccine. Understanding the mechanism adopted by remdesivir would add to the information on the drug-action mechanism. This would be of importance to the experimental and pharmaceutical labs in the process of drug development.
In your opinion, what are the key design considerations for your study?
The key design considerations of our study was to mimic the RNA dependent RNA polymerase inhibition by remdesivir. Trying to understand the structure of RdRP and designing the best fit molecule which can inhibit the drug target with more potency. Being a part of, one of the High Performance Computing (HPC) groups in India, the entire computational study was designed around making the best use of the HPC technologies available to us.
Which part of the work towards this paper proved to be most challenging?
The research work involved many challenges since we had to design simulation systems with limited and evolving structural information of this virus. The major challenge being performing the computational drug repurposing studies in order to accelerate this research. In addition, the analytics involving statistical techniques for the identification of crucial residues and subdomains of the viral protein RNA dependent RNA polymerase also proved to be challenging.
What aspect of your work are you most excited about at the moment?
The mechanism of action and crucial interacting residues predicted through the computational methods in our present study were observed to match the experimental structures that are being elucidated for the RNA dependent RNA polymerase of the SARS-CoV-2.
What is the next step? What work is planned?
The work in this article dealt with remdesivir action in inhibiting the RNA dependent RNA polymerase of SARS-CoV-2. Besides remdesivir, other nucleotide analogues are also known to inhibit RdRP from the other coronaviruses. Hence, we have planned to study the inhibitory mechanism of other nucleotide/nucleoside analogues namely, favipiravir, galidesivir, lamivudine, ribavirin and sofosbuvir in their active metabolite form. This work is currently being targeted using molecular docking and molecular dynamics simulations. We have planned to understand the conformational changes that the RdRP undergoes on binding to natural nucleotides and their analogues. This information may help in designing of better nucleotide analogues.
One more aspect we plan to study is the role of phytochemicals from medicinal plants that are known to be used as a treatment for respiratory ailments. All the data obtained through simulations has been thoroughly sampled and analyzed using statistically significant methods, such as, principal component analysis and Markov state modeling analysis.
Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2
Shruti Koulgi, Vinod Jani, Mallikarjunachari V. N. Uppuladinne, Uddhavesh Sonavane and Rajendra Joshi
RSC Adv., 2020,10, 26792-26803
DOI: 10.1039/D0RA04743K, Paper
 Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Keep up to date with our latest HOT articles, Reviews, Collections & more by following us on Twitter. You can also keep informed by signing up to our E-Alerts.