What can cause a mutation in DNA? Well, if you were to ask the Incredible Hulk (nicely), he would probably say– well not a lot, he’s more of a doer, but Bruce Banner might tell you gamma rays. But that is so 20th century.
In a Communication recently published in ChemComm, José Pedro Pedro Cerón-Carrasco (Université de Nantes) and Denis Jacquemin (Institut Universitaire de France) have shown that DNA can mutate permanently if an appropriate external electric field is applied.
Application of the right level of electric field can lead to proton transfer, which can cause the formation of tautomers, i.e. isomers of the DNA bases. By interfering with the bases and their interaction, a mismatch or mutation can be induced.
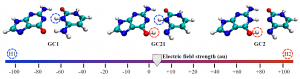
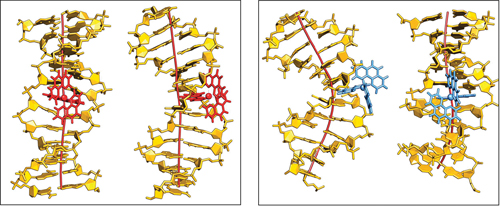
Turn the power up a little more and soon I will become Science Girl!: The DNA tautomers form under the influence of an external electric field. Circles indicate the protons that have been shifted compared to the canonical structure: H1 in blue and H2 in red.
Cerón-Carrasco and Jacquemin used a computational model to assess the effects of both positive and negative external electric fields on a DNA model to achieve an in vivo-like outcome. When applying an increasing strength of negative electric fields, they saw the more acidic H1 proton shift to the other base; intense positive fields activated the H2 proton.
The authors conclude that intense electric fields might damage DNA in a partially controlled way. This could have exciting applications for biochemistry or medicine– for example, selectively mutating a disease-causing cell. Or maybe, bestowing me with superpowers…
Interested in more? Read this HOT ChemComm article in full!
Electric field induced DNA damage: an open door for selective mutations
José Pedro Pedro Cerón-Carrasco and Denis Jacquemin
Chem. Commun., 2013, Accepted Manuscript
DOI: 10.1039/C3CC42593B
Sarah Brown is a guest web-writer for Chemical Communications. Sarah hung up her lab coat after finishing her PhD and post-doctorate in nanotechnology for diagnostics and therapeutics to become an assistant editor at the BMJ Publishing Group. When not trying to explain science through ridiculous analogies, you can often find her crocheting, baking and climbing, but not all at once.