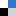A few years ago, when I discovered what an intercalator was, I thought it would be a great name for a burger bar (probably best situated near a chemistry department). In scientific terms (and not catering as sadly the idea didn’t take off), intercalators have attracted a great deal of attention and are best known for their use in anticancer treatments.
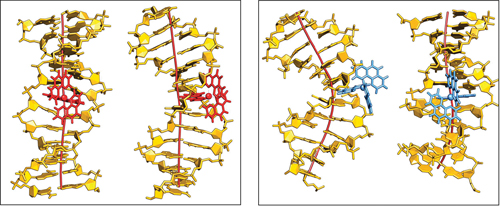
Chelate compounds of polycyclic heteroaromatics with transition metals can bind to DNA. The polycyclic moeities intercalate between the base pairs of the DNA, a little like the burger in a bun. This can have a profound effect on the DNA’s structure, separating the base pairs and causing the helix to kink. However, the extent of this effect is dependent on parameters such as the ligand and which enantiomer of the instrinsically chiral compounds is involved.
A study by Anna Reymer and Bengt Nordén into the ruthenium compound, [Ru(phenanthroline)3]2+, investigates its two enantiomers (Δ and Λ) and the effect each one has on binding specificity with DNA. Using molecular dynamics simulations, they demonstrated that the Δ-form induced a kink of 53° whilst the Λ-form produced a more typical bend of only 16°. They also reveal information about binding affinities of the compounds and how ‘deeply’ they can insert themselves into the base stack.
This interesting simulation is significant in the context of chiral recognition and evolutionary selection. It also gives further insight into the behaviour of DNA-protein interactions; an analogous kink as produced by Δ- [Ru(phenanthroline)3]2+ have been observed for several classes of operatory proteins.
To find out more download Reymer and Nordén’s communication.