Scientists have found that the way ice bonds to metal does not obey the ‘ice rules’. Andrew Hodgson, together with teams from the UK and Spain, wanted to understand water–metal and hydroxyl–metal interactions, to devise molecular models of wet metal interfaces for studying catalytic and electrochemical reactions that occur on these types of surfaces.
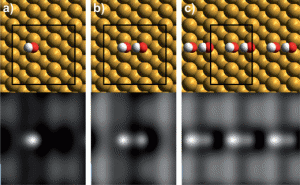
Using scanning tunnelling microscopy and density functional theory calculations, the teams produced a phase diagram for water and hydroxyl on a copper surface, providing a complete molecular description of the complex hydrogen bonding structures formed. They saw three distinct phases as the temperature was decreased and the water/hydroxyl ratio increased: pure OH dimers, extended 1H2O:1OH chains aligned along the close-packed Cu rows, and finally a distorted 2D hexagonal c(2 × 2) 2H2O:1OH network.
Binding geometry and simulated STM images for (a) an isolated OH group, (b) an OH dimer and (c) an array of OH forming a dimer chain on the copper surface
None of these phases obey the conventional ‘ice rules’. Instead, their structures can be understood based on weak H donation by hydroxyl, which favours H-bonding structures dominated by water donation to hydroxyl, and competition between hydroxyl adsorption sites.
Found out more by downloading the Chemical Science Edge article.