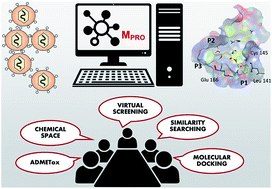
We are very pleased to introduce Marisa G. Santibáñez-Morán (first author), José Medina-Franco (corresponding author) and the team behind the paper ‘Consensus virtual screening of dark chemical matter and food chemicals uncover potential inhibitors of SARS-CoV-2 main protease‘. Their article has been very well received and handpicked by our reviewers and handling editors as one of our HOT articles. The team told us more about the work that went into this article and what they hope to achieve in the future. You can find out more about the authors and their research at DIFACQUIM, Computer-aided drug-design at UNAM, and find more HOT articles in our online collection.
Could you briefly explain the focus of your article to the non-specialist (in one or two sentences only) and why it is of current interest?
Our article looks for molecules in food chemicals or dark chemical matter (molecules that had not shown activity in 100 or more high-throughput screening assays) that are prospective inhibitors of the SARS-CoV-2 Main protease.
How big an impact could your results potentially have?
It could point to SARS-CoV-2 inhibitors that might otherwise have been overlooked. We would be glad if other research groups will be interested in the computational hits we made publicly available and further analyzed them in experimental assays.
Could you explain the motivation behind this study?
COVID-19 is currently affecting all aspects of human life. Our research group works on computer-aided drug design, and we had previously worked on drug repurposing. We felt that we could and should contribute to the collaborative efforts of scientists from all around the world.
In your opinion, what are the key design considerations for your study?
One was the selection of the molecular libraries where we looked for potential inhibitors. These comprise compounds that recent studies on SARS-CoV-2 have analyzed on a limited basis. Additionally, a large number of these molecules are ready to be tested in experimental assays. Moreover, there are currently numerous papers that reported favorable molecular docking results. However, selecting compounds that would have satisfactory potency and biopharmaceutical results in experimental settings is not trivial. Therefore, we ranked the compounds considering positive results by two molecular docking programs, Machine learning predictions, commercial availability, and ADMETox properties.
Which part of the work towards this paper proved to be most challenging?
First, to select a target and molecular queries for the structural similarity analyses. The latter should include structurally diverse and promising compounds. Another challenge was to create a classification method that helps us select compounds with better possibilities for drug development.
What aspect of your work are you most excited about at the moment?
I am excited about the possibility of finding supporting information about the activity of food chemicals against SARS-CoV-2. I believe that this could result in the development of nutraceuticals with inhibitory activity against the SARS-CoV-2 virus.
What is the next step? What work is planned?
We are waiting for the experimental results of 3 compounds that are being tested by our collaborators in North Carolina. We are also working on another manuscript that explores a broader region of the chemical space. And we hope that we could form new collaborations with RSC Advances readers.
Consensus virtual screening of dark chemical matter and food chemicals uncover potential inhibitors of SARS-CoV-2 main protease
Marisa G. Santibáñez-Morán, Edgar López-López, Fernando D. Prieto-Martínez, Norberto Sánchez-Cruz and José L. Medina-Franco
RSC Adv., 2020,10, 25089-25099
DOI: 10.1039/D0RA04922K, Paper
 Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Keep up to date with our latest HOT articles, Reviews, Collections & more by following us on Twitter. You can also keep informed by signing up to our E-Alerts.