We are very pleased to introduce Dr Shuntaro Takahashi and Professor Naoki Sugimoto, first author and corresponding author of the paper ‘Molecular crowding induces primer extension by RNA polymerase through base stacking beyond Watson–Crick rules‘. Their article has been very well received and handpicked by our reviewers and handling editors as one of our September HOT articles. The authors told us more about the work that went into this article and what they hope to achieve in the future. You can find out more about the authors and their article below and find more HOT articles in our online collection.
Meet the authors
 Shuntaro Takahashi is an Associate Professor at the Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University, Japan. Dr. Takahashi earned his PhD degree at Tokyo Institute of Technology in 2007. After a period of research at Tokyo Institute of Technology as an Assistant Professor, he joined FIBER in 2012. He is currently studying the biophysics of nucleic acids in cells and the mechanism of molecular crowding for nucleic acid structures that affect cellular metabolism.
Shuntaro Takahashi is an Associate Professor at the Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University, Japan. Dr. Takahashi earned his PhD degree at Tokyo Institute of Technology in 2007. After a period of research at Tokyo Institute of Technology as an Assistant Professor, he joined FIBER in 2012. He is currently studying the biophysics of nucleic acids in cells and the mechanism of molecular crowding for nucleic acid structures that affect cellular metabolism.
Professor Sugimoto received his PhD in 1985 from Kyoto University, Japan. After completing his postdoctoral work at the University of Rochester in the U.S.A., he became a faculty member at Konan University in Kobe, Japan in 1988. He has been a full professor since 1994 and a director at the Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University since 2003. He received The Imbach-Townsend Award from IS3NA in 2018. In 2020, he was awarded CSJ Awards from the Chemical Society of Japan. His research interests include biophysical chemistry, biomaterials, biofunctional chemistry, and biotechnology in the field of nucleic acid chemistry.
Could you briefly explain the focus of your article to the non-specialist (in one or two sentences only) and why it is of current interest?
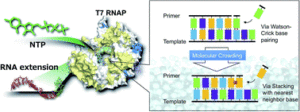
We investigated the effect of chemical environments on gene replication of the virus RNA polymerase. This article provides insight into not only the evolution of life but also the mechanism of mutation of the virus genome including SARS-CoV-2.
How big an impact could your results potentially have?
Our results provide one story that the molecular environment could take part in the evolution of life by enhancing the replication error of genome sequences. Moreover, this study suggests the significance of molecular environments of patients’ cells for spreading viruses.
Could you explain the motivation behind this study?
The stability of the Watson-Crick base pair is NOT always the most stable, which can be perturbed by molecular environments. Therefore, we speculated that the replication of nucleic acids in the enzyme could also be affected by molecular environments and cause replication errors.
In your opinion, what are the key design considerations for your study?
The key design consideration of our study is to quantitatively understand the effect of molecular environments on the replication fidelity because the stability of nucleic acids structures depends on the physicochemical properties of the solution such as dielectric constant and water activity.
Which part of the work towards this paper proved to be most challenging?
For the reagents for the molecular environments, we used poly(ethylene glycol)s. Although these reagents were easy to tune the solution properties, the effect on RNA and protein were different and complex. The choice of suitable condition was very important for this kind of research.
What aspect of your work are you most excited about at the moment?
We were excited to find the replication rules became dependent on the stacking interactions more than Watson-Crick base pairing under molecular crowding conditions. This indicates that the replication error can be simply explained by the changes in dielectric constant.
What is the next step? What work is planned?
This study suggests that the rule of the base pairings can be differentiated under molecular crowding conditions. Thus, we will pursue the biological role of non-Watson-Crick base pairings such as Hoogsteen base pairs under different cellular conditions. We are also interested in the effect of molecular environments on the reaction of RNA-dependent RNA polymerase of Covid-19.
Molecular crowding induces primer extension by RNA polymerase through base stacking beyond Watson–Crick rules
Shuntaro Takahashi, Hiromichi Okura, Pallavi Chilka, Saptarshi Ghosh and Naoki Sugimoto
RSC Adv., 2020,10, 33052-33058
DOI: 10.1039/D0RA06502A, Paper
 Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Submit to RSC Advances today! Check out our author guidelines for information on our article types or find out more about the advantages of publishing in a Royal Society of Chemistry journal.
Keep up to date with our latest HOT articles, Reviews, Collections & more by following us on Twitter. You can also keep informed by signing up to our E-Alerts.