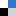Researchers in China have been able to visualize the intracellular location of DNA origami with a label-free fluorescent probe.
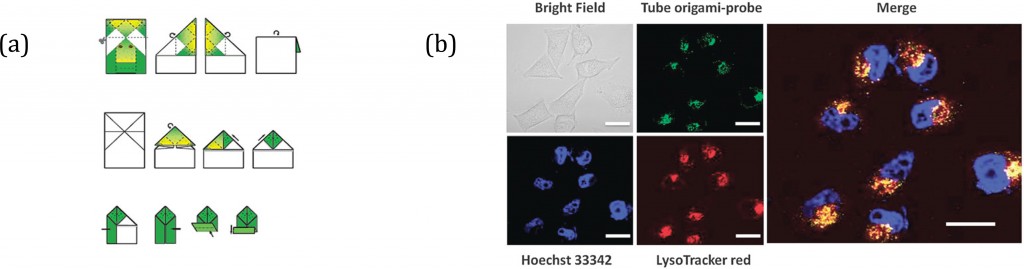
But let’s unfold a few things first and figure out what that means. DNA origami is the folding of a strand of DNA to make arbitrary 2 or 3 dimensional shapes; this serves as a ‘scaffold’ for shorter DNA strands that help hold the structure in its folded shape. These structures may be used for drug delivery, biosensors and more. I once made an origami frog; I wonder if there are any similarities…
Direct visualisation of the distribution and stability of DNA origami in live, cellular systems has not been achieved. Fluorescent labels can be attached to DNA strands but these have their drawbacks, such as weak emission intensity, photobleaching and expensive. Ding and co-workers looked at alternatives to visualize DNA origami in live cells.
The group were inspired by research on a series of carbazole-based cyanine fluorescence probes, which have a weak emission when they are monomolecularly dissolved but switch to a strong luminescent state upon binding to DNA or protein molecules. The significant enhancement is attributed to restricted intramolecular rotational (RIR) motions by anchoring the DNA molecules, which causes the large reduction in the non-radiative decay of fluorescence molecules.
Ding and co-workers then took some tubular DNA origami, the cyanine fluorophore and found that the carbazole-based cyanine molecules could be used as a sensitive optical switch, turned on when DNA origami is detected and turned off when the nanostructure degrades. After incorporating the cyanine probe molecules, the DNA origami-probe complex was administered to live cells. Excitingly, the green-yellow frog… erm, I mean, fluorescence was visible inside the cells treated with the probe. The group went further to try and understand the internalization mechanism of the DNA origami and found the probe localized in lysosomes. Finally, degradation studies showed that most DNA origami were dissociated after 60 hours, also a bit like my origami frog.
Unlike my attempts at origami, Ding and co-workers have demonstrated an exciting step in scaffolded DNA origami and its future applications in nanomedicine.
Read this HOT Chem Comm article today (free to access until the 5th of December 2012):
Visualization of the intracellular location and stability of DNA origami with a label-free fluorescent probe
Xibo Shen, Qiao Jiang, Jinye Wang, Luru Dai, Guozhang Zou, Zhen-Gang Wang, Wei-Qiang Chen, Wei Jiang and Baoquan Ding
Chem. Commun., 2012, 48, 11301-11303
Published on behalf of Sarah Brown, Chemical Communications web science writer.