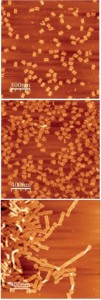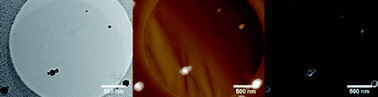
Swarming E. coli.
There are an estimated 1030 bacteria on Earth. The number of bacteria is greater than the number of stars in the Universe and is growing exponentially.
Bacteria are generally studied in the laboratory in Petri dishes under very well defined conditions. However, bacteria also thrive in more complex environments where the conditions are constantly varying. Some of these environmental changes are regular e.g. variations in light intensity from day to night, while others are random e.g. temperature, food availability and the presence of toxins or other bacteria.
Bacteria have developed a number of strategies to survive in these fluctuating environments. In the opening talk of the DPG spring meeting in Berlin last week, Stanislas Leibler from the Rockefeller University, New York and the Institute for Advance Studies, Princeton, discussed recent experimental and theoretical studies exploring the complex behaviour observed in bacterial colonies.
Consider a growing colony of bacteria. When an environmental change occurs one of two things may happen if the colony is to survive. (1) The bacteria ‘senses’ the change and changes to a state that is adapted for this new environment. This is known as responsive switching. (2) A small minority of the bacteria in the colony are poorly adapted to the initial environment. However, they become the most-adapted when the environment changes and survive while the rest are killed; the minority becomes the majority. This is known as stochastic switching.
So which is it? For colonies of bacteria with antibiotic persistence, experiments suggest that stochastic switching is the dominant behaviour. Leibler’s group added Ampicillin to growing colonies of Escherichia coli. The majority of the colony dies, but a few resistant bacteria survive. These resistant bacteria are able to grow, forming a new colony, once the antibiotic is removed. The persistent bacteria have a different phenotype to the rest of the colony. Under normal conditions, they grow much more slowly than the non-resistant bacteria, but are not killed when the antibiotics are added. Although the presence of these persistent cells leads to a lower population fitness, they act as an insurance policy and ensure that the colony can survive in the event of an antibiotic encounter. Leibler believes that this heterogeneity of bacterial populations is important for their ability to adapt to fluctuating environments and the persistence of bacterial infections.
While important when considering antibiotic resistant infections, these results may have much wider implications in areas ranging from cancer treatments, to models of financial investments, to information theory and statistical mechanics.
For more information see:
Balaban, N.Q. et al., Bacterial persistence as a phenotypic switch, Science, 2004.
Kussell, E. et al., Bacterial persistence: A model of survival in changing environments, Genetics, 2005.
Rivoire O, Leibler S, The value of information for populations in varying environments, J. Statist. Phys., 2011.
The image is taken from: Bacterial swarming: a model for studying dynamic self-assembly, Soft Matter, 2009, and shows a swarming colony of E. coli bacteria.
Comments Off on Survival in the face of the unknown