Molecules capable of mimicking the binding and/or functional sites of proteins represent a promising avenue for the development of potential drug candidates. This strategy allows for the incorporation of key structural features into simpler scaffolds and opens a wide range of opportunities for developing molecules with enhanced and modular biological activities.
A recent OBC publication by Professor Rob Liskamp of Glasgow University addresses a current challenge in the development and application of complementarity determining region (CDR) mimics, which have recently been shown to successfully behave as synthetic antibody mimics.
The concept of simplifying large proteins into simpler structures requires the synthesis of preorganized molecular scaffolds, which function as the core structural unit for attaching the biologically active CDR component. Such analogues have been shown to possess increased bioavailability, proteolytic stability and exhibit reduced immunogenic responses.
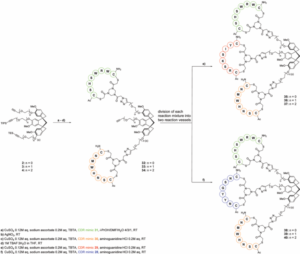
The group had previously reported the synthesis of a CTV-derived scaffold (Figure, compound 2), onto which different peptide segments could be incorporated, essentially generating synthetic CDRs with novel and tuneable physico-chemical properties. Poor solubulity however, has limited the progress of this class of compound in the drug discovery process.
Their current study focuses on the development of a scalable, one-pot synthesis of water soluble CTV-derived scaffolds (Figure, compounds 3, 4), which incorporate mono or diethylene glycol spacers. Late-stage diversification of the CTV-derived scaffolds is ammenable through Cu(I)-catalyzed azide-alkyne cycloaddition. This allows for the generation of a diverse series of synthetic antibodies, which were shown to mimic the antigen binding site of monoclonal antibody (mAb) infliximab (Remicade)—used for the treatment of human tumour necrosis factor alpha (hTNFa) mediated autoimmune diseases. SPR binding studies against the hTNFa receptor identified 5 leads with KD’s measured between 11 and 66 mM. While further modifications are required to improve solubility for evaluation in vitro, this study demonstrates the potential of this work to extend beyond antibody mimics. As any azide handle can be linked to the CTV-derived scaffold, one can envision the application of this methodology toward alternative protein mimics.
To find out more see:
Synthetic antibody protein mimics of infliximab by molecular scaffolding on novel CycloTriVeratrilene (CTV) derivatives
Ondřej Longin,
DOI:10.1039/C8OB01104D
Victoria Corless is currently completing her Ph.D. in organic chemistry with Prof. Andrei Yudin at the University of Toronto. Her research is centred on the synthesis of kinetically amphoteric building blocks which offer a versatile platform for the development of chemoselective transformations with particular emphasis on creating novel biologically active molecules.