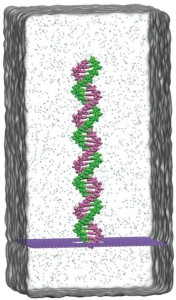
Quickly detecting DNA sequences with high accuracy is a significant goal for diagnostic medicine and genetics. The vast quantities of DNA and its small size make it difficult to achieve this goal. Nanopores offer a rapid method of detection by threading strands of DNA through a nanopore to detect individual nucleotides of the DNA. Solid-state nanopores separate two fluidic chambers and as the DNA passes through the pore, the pore becomes partially clogged and produces a blockage of ionic currents. The change in ionic current can be used to collect structural information from the DNA. Graphene has been well studied, experimentally and theoretically, for fabrication of these pores, but graphene produces significant complications with DNA detection. Graphene pores often contain material defects and DNA sticks to graphene, blocking the pores permanently, limiting detection capabilities. In a recent full article in Soft Matter, a collaborative research team tackles this issue by theoretically modeling DNA in a boron-nitride
Boron-nitride is composed of boron and nitride atoms in a honeycomb structure. The material is just as thin as graphene and exhibits similar desirable electrical and mechanical properties, but is more resistive to DNA adhesion, allowing the DNA to pass through the pore without permanent blockage. The research group used large-scale molecular dynamics to model double stranded DNA passing through boron-nitride pores ranging in size from 2.5 to 6.5 nm with an external voltage of 1.0 V. The smaller 2.5 nm pore size had greater blockage currents and a higher sensitivity to DNA threading through the pore than the larger pores due to its smaller cross-sectional area. Double stranded DNA composed of only adenine-thymine (A-T) or guanine-cytosine (G-C) nucleotide pairs were compared using the 2.5 nm pores with multiple applied voltages with the goal of understating the changes is current signal as the DNA passed through the pore. The greatest difference between A-T DNA and G-C DNA was observed at 1.0 V with G-C DNA exhibiting greater stretching and stress than the A-T DNA. Both sets of DNA passed readily through the boron-nitride nanopores without permanent blockage. In comparison, in modelling of DNA passing through graphene nanopores, DNA exhibited significant adhesion and breakage of the DNA at the pore. The large changes is current signal between A-T and G-C DNA and the lack of DNA adhesion using boron-nitride pores offer an exciting opportunity for future DNA sequencing. The molecular dynamic modeling presented will hopefully influence current experimental work in the development of DNA detection with nanopores.
See the full Soft Matter article here:
DNA translocation through single-layer boron nitride nanopores
Zonglin Gu, Yuanzhao Zhang, Binquan Luan and Ruhong Zhou
a
Dr. Morgan M. Stanton is currently a postdoctoral researcher at the Max Planck Institute for Intelligent Systems in Stuttgart, Germany. She completed her Ph.D. in Chemistry from Worcester Polytechnic Institute in 2014. Read more about Morgan’s research publications here or you can follow her on Twitter @morg368.
Follow the latest Soft Matter publications and updates on Twitter @softmatter or on Facebook