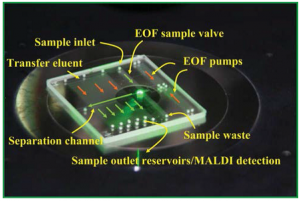
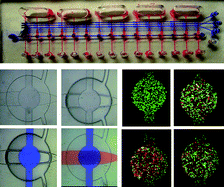
Researchers at Virginia Tech have designed a microfluidic system that integrates optimized fluid handling, liquid chromatography, and a mass spectrometry sample platform – all in one small device.
Matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) is a powerful analytical tool which enables the identification and quantification of thousands of proteins and peptides within a single complex sample. Liquid chromatography (LC) can be used to pre-sort sample contents by mass, increasing the sensitivity and selectivity of the MS measurements. A novel system designed by Iulia Lazar and Jarod Kabulski (view the full paper here) allows the entire process of LC-MS to be carried out on a microfluidic chip.
Lazar and Kabulski’s chip has several features that set it apart from previous microfluidic devices incorporating LC and MS.1 It comprises a unique microfluidic system2 driven by electro-osmotic pumps, an LC channel packed with microparticles, and a novel system of transverse microchannels to draw LC fractions from the main separation channel and into a series of reservoirs. The chip can then be directly loaded into the MALDI-MS instrument for analysis. Flow control in the LC channel was carefully optimized so that the solution components would be perfectly distributed along the channel length.
Using the microfluidic LC-MS chip, the researchers were able to obtain results comparable to conventional LC-MS, for both peptide mixtures and cytoplasmic cell extract. Thus, the chips are very promising for practical high-throughput applications such as biomarker screening. The chip format will also enable samples to be collected and prepped by an MS non-expert at a remote location, then transported to a MS lab for analysis.
Learn more from this HOT article from Lab on a Chip!
Microfluidic LC device with orthogonal sample extraction for on-chip MALDI-MS detection
Iulia M. Lazar and Jarod L. Kabulski
DOI: 10.1039/c3lc50190f
- D. Gao et al., Lab on a Chip 13, 3309-3322, 2013.
- I. Lazar et al., Analytical Chemistry 78 (15), 5513-5524, 2006.