In a recent paper in Lab on a Chip, which featured as the Cover Article of Issue 22, Vol 13, researchers from the Ecole Polytechnique, Palaiseau, France describe a micro-droplet based method for measuring reaction kinetics using tiny (20 nL) sample volumes.
When working with precious reagents such as enzymes, minimizing sample volume is of the utmost importance. Droplet-based microfluidic methods can greatly reduce sample volumes, but still waste considerable amounts of sample in fluid handling processes (e.g. pushing the sample through a channel). A team led by Marten Vos and Charles Baroud has utilized a unique “rails and anchors” method1 of forming and controlling the movement of droplets to observe the kinetics of a chemical reaction using only 20 nL of each reagent solution.
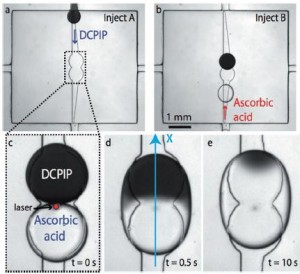
In this scheme, the flow chamber geometry is carefully designed so that when an aqueous solution is injected into the oil-filled chamber, droplets of a reproducible size are released and constrained between the top and bottom surfaces of the chamber. Then, widening grooves (“rails”) in the bottom of the chamber guide the droplets via surface tension to “anchors” in the center. (See figure a and b.) In this way, no additional solution is wasted pushing droplets around.
The authors used the “rails and anchors” to place two droplets, each containing a different reagent, in proximity, and then used an IR laser pulse to break the surface tension between them (figure c). The interface between the two solutions (in this case, DCPIP and ascorbic acid) can clearly be seen in figure d below. By tracking the front of the color change that moves across the droplet as the reaction proceeds (figure e), it is possible to calculate the reaction rate (accounting also for the diffusion rates of the reactants).
The researchers also demonstrated the potential of multiplexing their technique by monitoring six different reactions in parallel droplets. They were able to obtain results in good agreement with a commercial stopped-flow spectrometer with greatly reduced cost and time.
Read this HOT article in Lab on a Chip today!
Parallel Measurements of Reaction Kinetics using Ultralow Volumes, Etienne Fradet, Paul Abbyad, Marten H. Vos, and Charles N. Baroud, DOI: 0.1039/c3lc50768h
References:
- P. Abbyad et al., Lab on a Chip 2011, 11, 813-821.