The study and optimisation of microalgal growth is a hot topic at the moment due to the use of microalgae in many industrial processes, as well as its potential use as biofuel. Previously, I have written about a Lab on a Chip article from the Sinton lab on optimising microalgal growth by varying irradiance conditions.
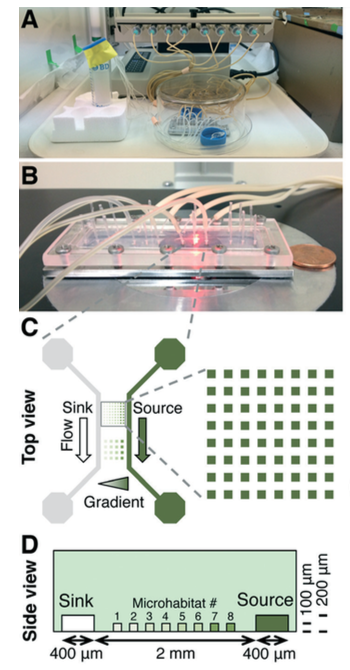
Now Mingming Wu’s group, from Cornell University, have published an article focused on the effect of nitrogen concentration on cell growth rates. Wu has developed a platform based on agarose gel, as shown in the diagram. The nutrient media can flow through this gel while the cells can’t, maintaining separate microhabitats.
The authors decided to study the effect of nitrogen concentration gradients on the microalgae (C. reinhardtii), using ammonium as the nitrogen source. Nitrogen is essential for microalgae, as it is required for protein and nucleic acid synthesis, and ammonium is the preferred source for this particular strain.
An ammonium gradient was obtained by flowing ammonium-containing media through the source channel, and ammonium-free media through the sink channel (diagram C). As expected, increasing the concentration (within the micromolar range) increased the microalgal growth rates. Fluorescence imaging allowed the authors to quantify the growth kinetics using the Monod equation (similar to the Michaelis-Menten equation for enzyme kinetics). This is the first time this has been achieved for this particular microalgal strain with nitrogen concentration as the variable.
Another interesting find was that when the microalgae were subjected to millimolar ammonium concentrations, growth inhibition was seen. The standard medium for microalgae contains 7.5 mM ammonium, so these results suggest that these concentrations need to be reduced by several orders of magnitude in order to maximise growth rates!
Wu and co-workers have nicely demonstrated the capablilty of their agarose-based platform in quantifying growth kinetics and they highlight that it is 50-fold faster, and more cost effective, than the standard chemostat system. They also observed cell heterogeneity during their experiments and plan to use their system to study this further, along with other aspects of cellular behaviour such as quorum sensing.
To download the full article for free* click the link below:
An array microhabitat system for high throughput studies of microalgal growth under controlled nutrient gradients
Beum Jun Kim, Lubna V. Richter, Nicholas Hatter, Chih-kuan Tung, Beth A. Ahner and Mingming Wu
Lab Chip, 2015,15, 3687-3694
DOI: 10.1039/ C5LC00727E
—————-

About the webwriter
Claire Weston is a PhD student in the Fuchter Group, at Imperial College London. Her work is focused on developing novel photoswitches and photoswitchable inhibitors.
—————-
*Access is free until 19/10/2015 through a registered RSC account – click here to register