For more than a century, researchers have been studying how the body detects and defends itself against foreign invaders.1 This process relies on a series of cell-to-cell signaling, starting with the detection of common pathogen components by receptors of immune cells. For example, macrophages, a surveillance immune cell, recognize bacteria using the Toll-Like receptor 4 (TLR4), which binds to lipopolysaccharide (LPS) found in the membrane of gram-negative bacteria.
Upon recognition of danger signals, immune cells engulf and degrade the invaders and activate NF-κB, an intracellular protein that, when activated, migrates into the nucleus and turns on inflammatory response genes. For immune cells, this initiates the secretion of signaling mediators such as tumour necrosis factor (TNF), which helps remove invaders by dilating local blood vessels and recruiting other immune cells.
Although our understanding of the immune system has skyrocketed in the past two decades,2 researchers are still lacking the tools needed to study the signaling of inflammation in a highly controlled manner. Most approaches rely on bulk population measurements, in which important spatial and temporal patterns of signaling are obscured by biological noise from neighbouring cells. To address this challenge, Tino Frank and Prof. Savas Tay from ETH Zurich developed a microfluidic system that can propagate inflammatory signals from a single immune cell (sender cell) and to a population of fibroblast cells (receiver cells).3
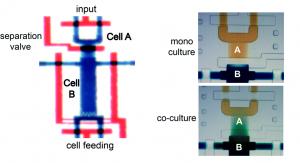
The microfluidic device, formed from polydimethylsiloxane (PDMS), comprises a deflectable membrane sandwiched between a flow layer and a control layer. The flow layer houses supply channels and cell culture chambers, while the control layer regulates input, output, and segregation of these chambers using solenoid-actuated valves.
As shown in picture 1, the chambers for the sender cell (chamber A) and receiver cells (chamber B) can be segregated for mono culture or connected for co-culture, depending on the state of the reversible separation valve. In mono culture mode, the sender cell can be stimulated independent of the receiver cells, whereas in co-culture mode, the signaling mediators secreted by the sender cell can be broadcasted to the receiver cells via diffusion.
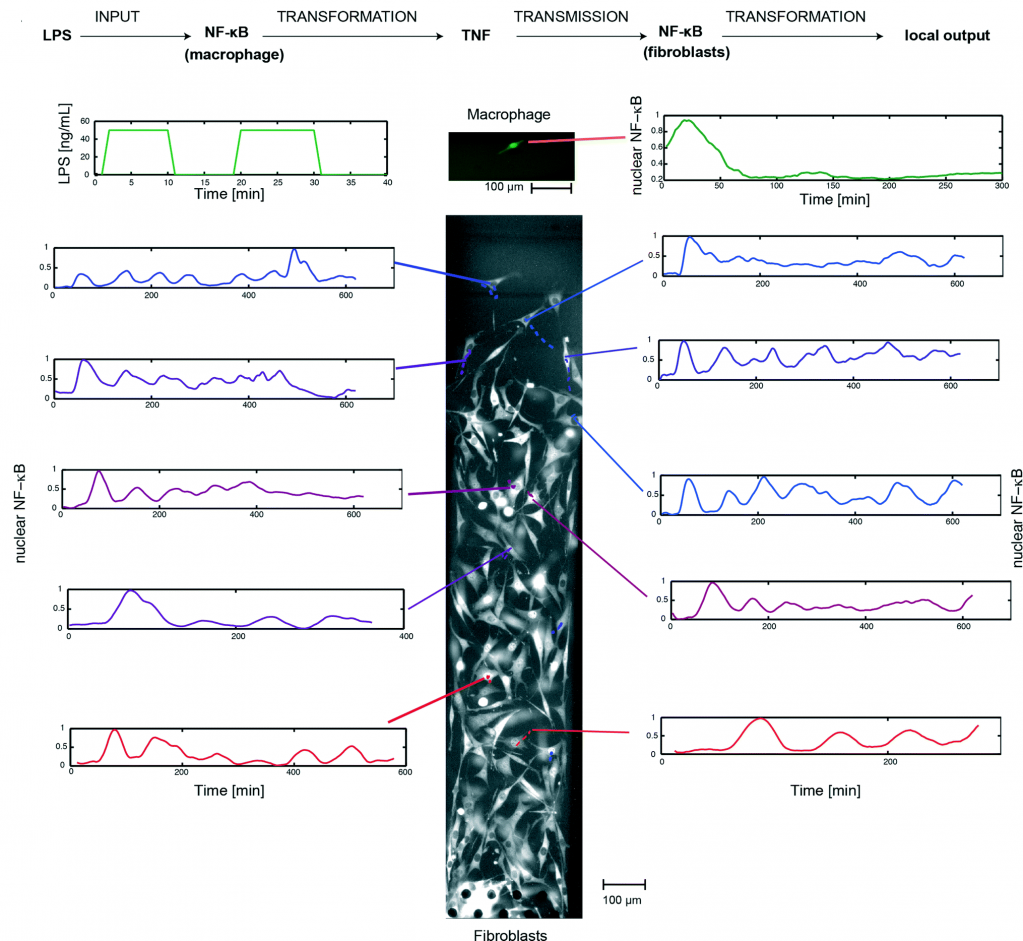
Using this system, the researchers implemented a single-cell model of inflammation arising from bacterial infection (picture 2). This model begins in mono culture mode, with a single macrophage seeded in chamber A and a population fibroblasts seeded in chamber B. Inflammation was initiated by stimulating the macrophage with short pulses of bacterial LPS, leading to the activation of NF-κB and TNF secretion. Subsequently, the device was switched to co-culture mode, allowing the secreted TNF to diffuse across the fibroblasts and activate their NF-κB signaling.
By tracking the movement of NF-κB in the nucleus of individual cells, the researchers, for the first time, mapped the spatiotemporal distribution of immune response to bacterial infection. They demonstrate that a single macrophage can activate and control over 100 fibroblasts up to 1 mm away, and that fibroblasts located farther away exhibited a time-delayed activation profile and fewer NF-κB oscillations (between the nucleus and cytoplasm). Furthermore, they observed that the NF-κB signaling in some fibroblasts can linger for up to 10 hours, demonstrating that a brief exposure to pathogenic signal can induce long-term inflammatory response in nearby cells.
In summary, Tino Frank and Prof. Savas Tay developed a microfluidic platform to study spatiotemporal dynamics of immune response in single cells. The continued investigation of these dynamics will be important for understanding of the inflammatory process and modeling of relevant signaling pathways.4
To download the full article for free* click the link below:
Automated co-culture system for spatiotemporal analysis of cell-to-cell communication
Tino Franka and Savaş Taya
Lab Chip, 2015, 15, 2192-2200
DOI: 10.1039/C5LC00182J
—————-
References:
1. K. M. Murphy, Janeway’s immunobiology, Garland Science, 2011.
2. L. A. J. O’Neill, D. Golenbock and A. G. Bowie, Nat Rev Immunol, 2013, 13, 453-460.
3. T. Frank and S. Tay, Lab on a Chip, 2015, 15, 2192-2200.
4. M. Junkin and S. Tay, Lab on a Chip, 2014, 14, 1246-1260.
—————-

About the webwriter
Alphonsus Ng is a postdoctoral fellow under the supervision of Dr. Aaron Wheeler in the Department of Chemistry and the Institute of Biomaterials and Biomedical Engineering at the University of Toronto. His research focuses on the development of microfluidic methods for heterogeneous immunoassays, cell-based assays, enzymatic catalysis, sample preparation for proteomics, and chemical synthesis.
—————-
*Access is free until 24/08/2015 through a registered RSC account.