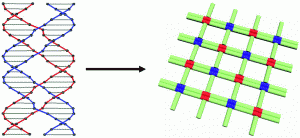
Nature uses a cooperative interplay of noncovalent interactions to control complex self-assembly of organized nanostructures with extreme precision. Taking this as inspiration, the field of structural DNA nanotechnology has been working toward the development of programmable, self-assembling nanomaterials and motion control at the nanoscale level by exploiting the remarkable molecular recognition properties of DNA. A number of basic structural motifs using branched DNA have been designed and are key elements for the construction of supramolecular arrays, molecular scaffolds and mechanical and logical nanodevices.
The paranemic crossover (PX) motif, as described in a recent OBC publication, has been of particular interest due to its unique ability to behave as both a self-assembling building block or tile and cohesive linker. Its application, until now, has been limited to 1D arrays as the level of flexibility within PX tiles needed to be controlled in order to access well-defined 2D and 3D DNA structures.
PX DNA arises from the fusion of two parallel double helices through strand cross-overs wherever the two strands come in contact. The component double strands are not linked and can hypothetically pair with each other indefinitely in a manner similar to the pairing of a buldged double helix. Advantageously, the PX structural motif reduces the need for sticky end cohesion, traditionally used in DNA-based self-assembly, which leads to an overall increase in nanostructure stability as sticky ends are susceptible to enzymatic degradation. In addition, complex DNA nanostructures made solely from PX motifs reduce topological problems during self-assembly thus minimizing error.
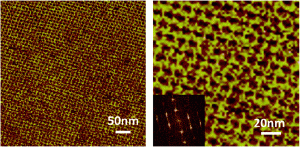
Researchers from the National Centre for NanoScience and Technology in Beijing, Anhui Normal University in China and Purdue University in the United States have collaboratively studied various structural parameters in order to optimize PX DNA’s ability to participate in the assembly of highly desirable 2D nanostructures. By varying the number of base pairs that make up the major (wide, W) and minor (narrow, N) grooves of the bulged double helix, several versions of PX tiles were prepared. Optimal parameters were observed when T65 (according to the formula TWN), which assembled into an extended, flat and regular 2D array (see image). Any deviation from this number of base pairs resulted in the tile becomes stressed and twisted leading to random aggregates.
This discovery has made possible the application of the highly desirable PX motif in 2D nanoconstruction which will no doubt lead to the synthesis of more stable and structurally and functionally intricate DNA self-assembling nanostructures.
To find out more see:
The study of the paranemic crossover (PX) motif in the context of self-assembly of DNA 2D crystals
Weili Shen, Qing Liu, Baoquan Ding, Zhiyong Shen, Changqing Zhu and Chendge Mao
DOI: 10.1039/c6ob01146b
Victoria Corless is currently completing her Ph.D. in organic chemistry with Prof. Andrei Yudin at The University of Toronto. Her research is centred on the synthesis of kinetically amphoteric molecules, which offer a versatile platform for the development of chemoselective transformations with particular emphasis on creating novel biologically active molecules.