 In their search to solve complex biological problems by bridging gaps between protein synthesis and biological application, Prof. Jennifer Ottesen and her group at Ohio State University have been successfully developing what they term a ‘chemical toolbox’ for histone protein synthesis.
In their search to solve complex biological problems by bridging gaps between protein synthesis and biological application, Prof. Jennifer Ottesen and her group at Ohio State University have been successfully developing what they term a ‘chemical toolbox’ for histone protein synthesis.
Within the field of epigenetics, heritable changes in gene expression outside of the DNA sequence are tightly regulated by post-translational modifications (PTMs) of DNA and histone proteins–the proteins that package DNA. Known PTMs of histone proteins include methylation, acetlyation, phosphorylation, sulfonylation and ubiquitination and fall under a hypothesized “histone code” which suggests that combinations of these markers alter DNA accessibility through chromatin restructuring and ultimately regulate gene expression.
In building synthetic histone proteins with distinct combinations of chemical modifications, the role of a specific sequence of PTMs in gene expression and the molecular mechanisms by which they function can be elucidated and targeted. This is of particular interest as epigenetics has become a hot topic in recent years due to an ever-growing understanding of these markers and their potential to act as selective entry points for disease intervention.
Ottesen’s recent publication in Organic and Biomolecular Chemistry outlines a new hybrid phase ligation approach for the synthesis of modified histone proteins which overcomes some long standing issues inherent in histone total synthesis. This method combines both solid and solution-phase ligation chemistry to improve process efficiency and overall yield. The group even demonstrates its ability to produce previously challenging CpA-K12ac histone protein which could not be synthesized with standard approaches.
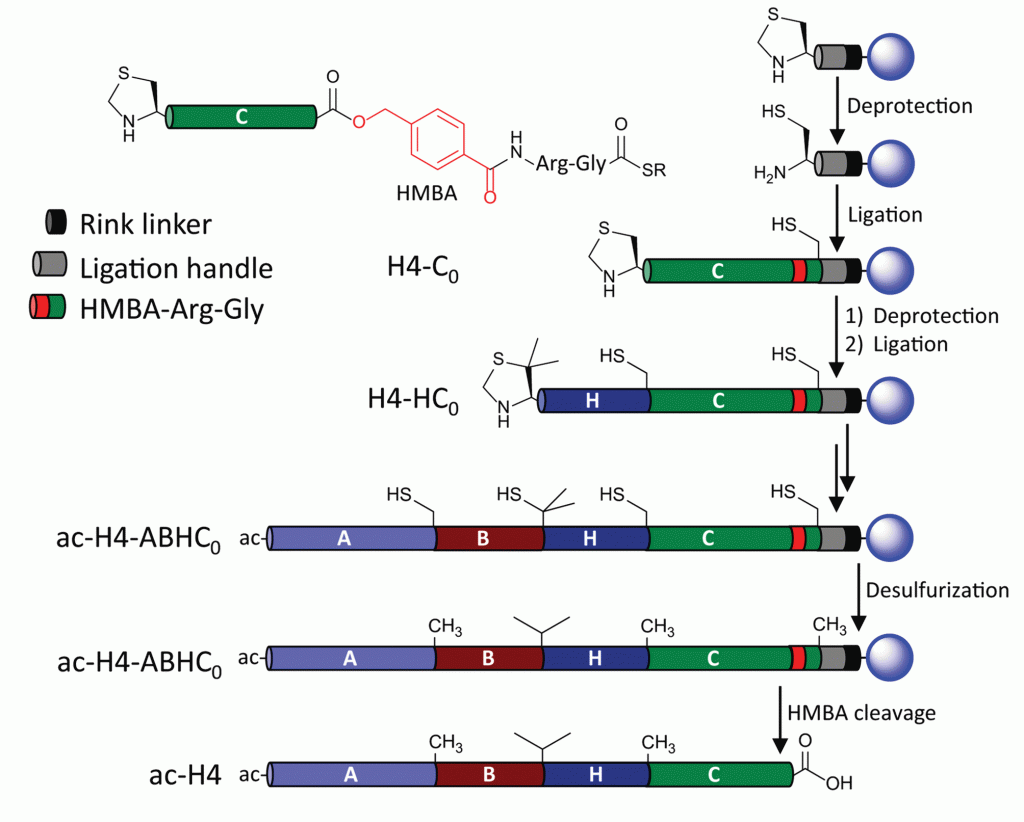 Key to their success is the application of a dual-linker strategy which led to an efficient, sequence-independent resin attachment that liberates the desired native carboxy terminus of the protein which had been previously difficult to accomplish. Below is a scheme describing the solid-phase native chemical ligation of one of their desired targets, histone H4. A single coupling cycle includes deprotection followed by ligation and cleavage from the resin may be accomplished at either the Rink linker (black), or at the HMBA linker (red) to generate the native terminus.
Key to their success is the application of a dual-linker strategy which led to an efficient, sequence-independent resin attachment that liberates the desired native carboxy terminus of the protein which had been previously difficult to accomplish. Below is a scheme describing the solid-phase native chemical ligation of one of their desired targets, histone H4. A single coupling cycle includes deprotection followed by ligation and cleavage from the resin may be accomplished at either the Rink linker (black), or at the HMBA linker (red) to generate the native terminus.
Studies such as Prof. Ottesen’s are crucial as mechanisms by which certain genes are regulated must first be determined before developing targeted therapeutic approaches. Histone deacetylase (HDAC) inhibitors for example, interfere with histone deacetylase and have shown activity against various cancers, neurological diseases and immune disorders. The utility of this class of compound depends on their ability to target and modulate a subset of genes without causing global biological changes. Presently, additional work is required to define the human epigenome, its role in disease development and the processes that regulate it. Progress in the synthesis of highly desirable modified histone proteins brings us ever closer.
To find out more see:
Hybrid phase ligation for efficient synthesis of histone proteins
Ruixuan R. Yu, Santosh K. Mahto, Kurt Justus, Mallory M. Alexander, Cecil J. Howard, and Jennifer J. Ottesen
DOI: 10.1039/C5OB02195B
Victoria Corless is currently completing her Ph.D. in organic chemistry with Prof. Andrei Yudin at The University of Toronto. Her research is centred on the synthesis of kinetically amphoteric molecules, which offer a versatile platform for the development of chemoselective transformations with particular emphasis on creating novel biologically active molecules.