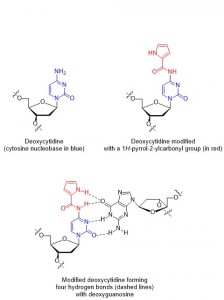
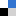Deoxyribonucleic acid (DNA) is one of the most famous molecules in the world. First isolated and identified by Freidrich Miescher in 1871,1 DNA is responsible for coding ‘genetic instructions’ in all known living organisms and viruses. DNA molecules consist of two long helical biopolymer chains wrapped around a common axis and held tightly together by intermolecular forces called hydrogen bonds and base-stacking interactions. Each biopolymer chain is made up of small units called nucleotides, which themselves consist of a sugar, a phosphate group, and one of four different nucleobases (guanine, adenine, thymine or cytosine, abbreviated to G, A, T and C respectively). The specific orders in which the nucleotides appear in a DNA strand is what allows for the storage of genetic information.
Shorter, single strands of nucleotide residues bonded together are called oligonucleotides. They are capable of binding to another oligonucleotide strand (via the same intermolecular forces as found in full length DNA helices) if the other strand has the ‘complimentary’ order of nucleotides (cytosine and guanine exclusively bind with each other; as do adenine and thymine). Due to this property, they are used in areas such as genetic testing, gene therapy, DNA probes and forensics.
The binding of one strand to its complimentary strand can be strengthened by modifying the nucleobase, as long as the modifications do not disrupt duplex formation. In particular, the nucleobase cytosine has several reported modifications2 which allow for additional hydrogen bonding (thus strengthening the binding of two strands). Unfortunately, a lot of the previously reported cytosine modifications have the drawback of long synthetic sequences necessary to make them.
In this paper, Sekine and co-workers report their modifications of cytosine nucleotides, and measure these new molecule’s binding affinities with guanine (the ‘complimentary’ base which cytosine hydrogen bonds with, Figure 1). As well as demonstrating an efficient synthetic route to their new cytosine derivatives, the authours prove that oligonucleotides which incorporate their modified cytosine residue show an increased binding affinity to the complimentary strands when compared with the unmodified, parent oligonucleotide. These promising results could lead to the design and synthesis of even better cytosine nucleotides inspired by the scaffold reported in this work, in turn leading to oligonucleotides which can perform better in DNA-recognition based tests.
To read more, see;
A new modified cytosine base capable of base pairing with guanidine using four hydrogen bonds
K. Yamada, Y. Masaki, H. Tsunoda, A. Ohkubo, K. Seio and M. Sekine,
Org. Biomol. Chem., 2014, DOI:10.1039/c3ob42420k. Download PDF
Free to access until 10th April
References
1 R. Dahm, “Discovering DNA: Friedrich Miescher and the early years of nucleic acid research” Human Genetics, 2008, 122, 565–81.
2 A. S. Wahba, A. Esmaeili, M. J. Damha, R. H. E. Hudson, Nucleic Acids Res., 2010, 38, 1048; S. Preus, K. Kilsa, L. M. Wilhelmsson, B. Albinsson, Phys. Chem. Chem. Phys., 2010, 12, 8881; A. Ohkubo, T. Sakaue, H. Tsunoda, K. Seio, M. Sekine, Chem. Lett., 2010, 39, 726.
 Dr C. Liana Allen is currently a post-doctoral research associate in the group of Professor Scott Miller at Yale University, where she works on controlling the enantio- or regioselectivity of reactions using small peptide catalysts. Liana received her Ph.D. in organic chemistry at Bath University with Professor Jonathan Williams, where she worked on developing novel, efficient syntheses of amide bonds.
Dr C. Liana Allen is currently a post-doctoral research associate in the group of Professor Scott Miller at Yale University, where she works on controlling the enantio- or regioselectivity of reactions using small peptide catalysts. Liana received her Ph.D. in organic chemistry at Bath University with Professor Jonathan Williams, where she worked on developing novel, efficient syntheses of amide bonds.