The 22nd International Conference on Miniaturized Systems for Chemistry and Life Sciences (aka MicroTAS) was held last year in Kaohsiung, Taiwan. Welcoming more than 1000 participants, MicroTAS 2018 conference brought together several disciplines including microfluidics, microfabrication, nanotechnology, integration, materials& surfaces, analysis & synthesis, and detection technologies for life sciences & chemistry. Besides the exciting scientific program and great presentations, social/networking events (welcome reception, student mixer, women night out, and conference banquet) have made MicroTAS 2018 conference an unforgettable one. In this article, we would like to share some of the conference highlights with our Lab on a Chip blog readers.
Unraveling endothelial cell phenotypic regulation by spatial hemodynamic flows with microfluidics
Sarvesh Varma, Guillermo Garcia-Cardena, & Joel Voldman
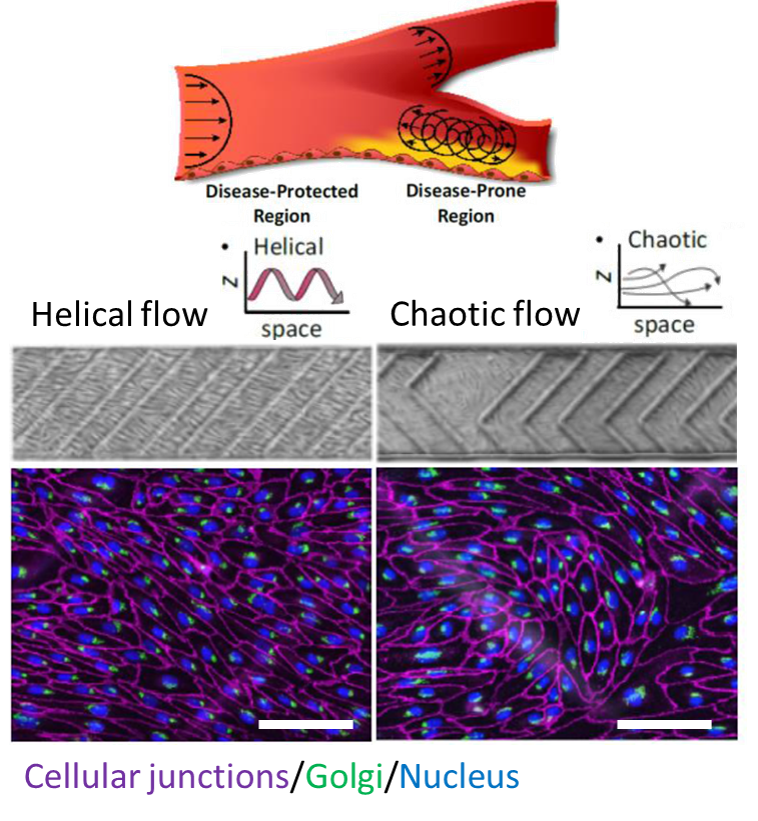
Did you know that artery bifurcations are prone to atherosclerosis? Blood flow profiles in vessels can help us to gain insights towards atherosclerosis. In this work, the authors fabricated a soft microdevice to study the effects of helical and chaotic flows on endothelial cells located in vein walls. They hypothesized and demonstrated that a helical (uniform) flow profile results in endothelial cells aligning upstream to flow and gain atheroprotective properties, while the chaotic flow results in misalignment of cells that give rise to atherosclerosis.
The figure shows the morphological adaptations of cells in response to distinct spatial flows, scale bars are 0.1 mm.
3D printing of microfluidic glass reactors
Patrick Risch, Frederik Kotz, Dorothea Helmer & Bastian Rapp
Microfluidic devices are mostly made from PDMS, although this material is not always well-suited for thermal, optical, mechanical and chemical changes. In this work, the authors present a new resin formulation to inspire the 3D printing of glass, which is more durable than PDMS. The resin was fabricated using stereolithography printer and this technique is useful for rapid prototyping of microfluidic devices made from glass for optical detection or chemical reaction applications.
A 3D gradient generator is shown in this figure, scale bar is 2 mm.
A Tetris-like modular microfluidic platform for mimicking multi-organ interactions
Louis Ong Jun Ye, Terry Chng, Chong Lor Huai, Seep Li Huan & Toh Yi-Chin
Modularization is undoubtedly on the rise in microfluidics and this work demonstrates an interesting approach. The authors focused on solving ‘limited compatibility with existing devices‘ problem. To achieve that, ring magnets were utilized to connect different parts of PDMS building blocks that were previously fabricated using micro molds. A modular platform assembled using this approach was shown to culture cells as a proof-of-concept study. The platform is expected to allow facile configuration of complex experimental set-ups involving multiple tissues.
The image shows a modular device (left), and its parts (right) connected each other via magnets, scale bars are 1 cm.

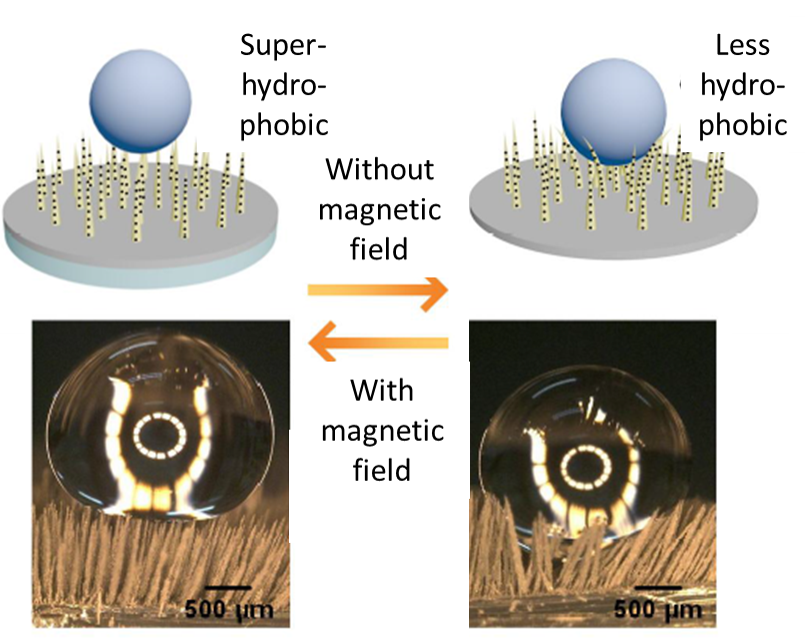
A magneto-switchable superhydrophobic surface for droplet manipulation
Chao Yang & Gang Li
Surface hydrophobicity is an important feature when it comes to bio and chemical applications. In this work, magneto switchable micro-pillars were made from PDMS and carbonyl iron particles. The pillars erect under influence of a magnetic field, resulting in subsequent switching of the wettability and adhesion of the surface between the water-repellent and water-adhesive states. The surface becomes superhydrophobic (water-repellent) when the magnetic field is applied. The authors demonstrated droplet lifting and transportation on a surface using this approach.
The image depicts the effect of an external magnetic field on the stiffness of micro-pillars.
About the Web writer
Burcu Gumuscu is a postdoctoral fellow in Herr Lab at UC Berkeley in the United States. Her research interests include the development of microfluidic devices for quantitative analysis of proteins from single-cells, next-generation sequencing, compartmentalized organ-on-chip studies, and desalination of water on the microscale.
Comments Off on MicroTAS 2018 Highlights