A new 3D printer and resin formula enable 3D printing of channels on the microscale
There is no doubt that 3D printing is a hot technology with applications across all sectors of industry and life. The technology is attractive not only for its use in rapid prototyping, but also for its ability to form complex structures that could not easily be formed by other manufacturing techniques. There is also the added benefit of digital design—it is very easy to go from a CAD model to physical production without the need of additional tooling or equipment. For these reasons and others, 3D printing is an attractive manufacturing process for microfluidics.
Albert Folch has argued that 3D printing is the solution to the barriers preventing the translation of PDMS microfluidics to commercial applications. (Angew. Chem. Int. Ed. 2016, 55, 3862–3881.) However, printing resolution has limited the size of channels to the milli- and sub-millifluidic regimes. The challenge when it comes to microchannels is that commercial 3D printers are designed to form polymer features but microchannels, by their nature, are voids within bulk material. While many 3D printer manufacturers tout resolutions in the tens of micrometers, the reality is that the resolution of the void spaces can be several-fold larger. In their recent report, researchers at Brigham Young University have advanced 3D printing technology to print channels truly in the micro- regime.

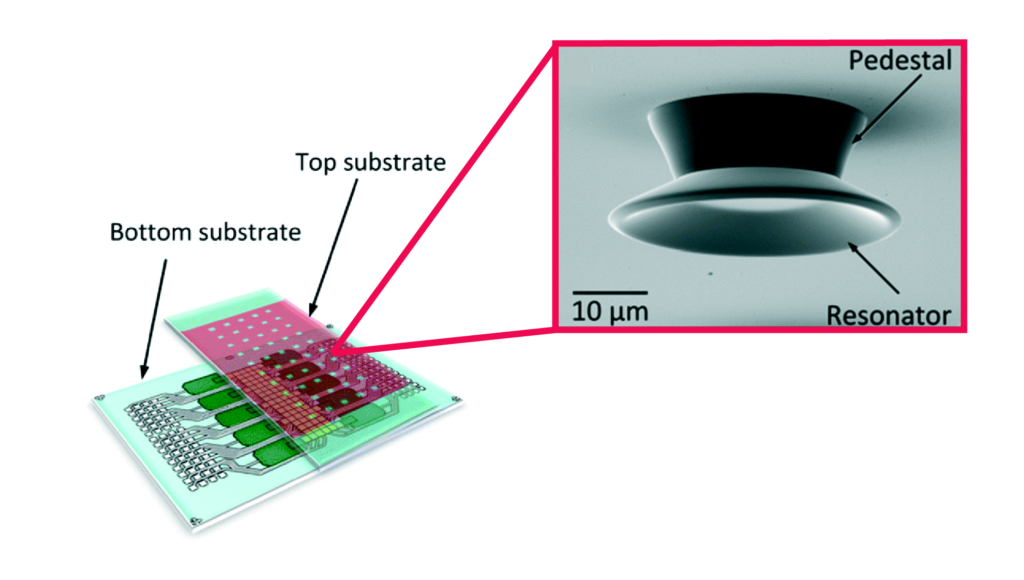
Left: Schematic illustration of 3D stacked serpentine channel design. Right: SEM image of 3D serpentine channel cross section.
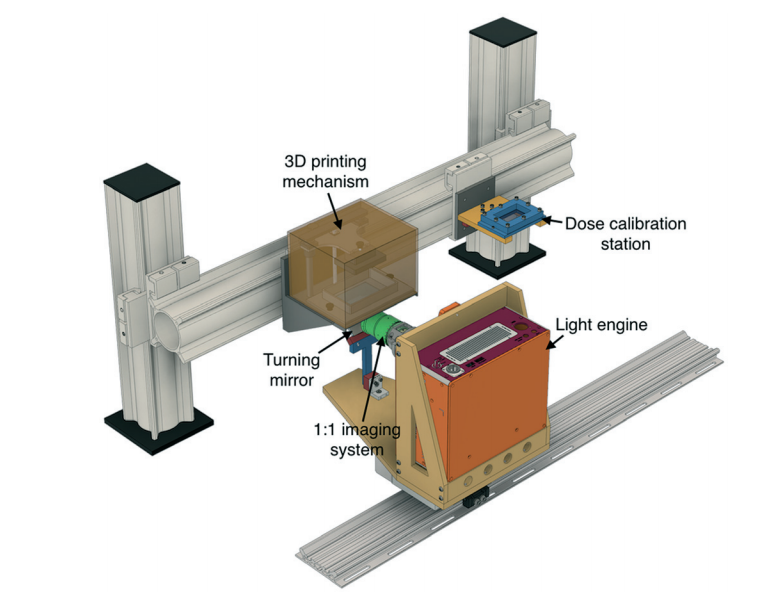
Achieving true micro-scale printing resolution required innovation in both the 3D printer and the resin material. Gong et al. modified a digital light processing (DLP)-type 3D printer to include a high resolution light engine and a 385 nm UV LED light source. DLP printers project a pattern of light over the entire plane, crosslinking an entire layer in a single step. The light engine enables resolutions in the x-y plane of 7.6 µm and their choice of a 385 nm light source opened a wide range of possible UV-absorbers to them. Twenty different UV-absorbers were evaluated and 2-nitrophenyl phenyl sulfide (NPS) was chosen as having all the desired properties, with the main criterion of enabling smaller void sizes in the z-dimension. The final composition of the resin was a mixture of poly(ethylene glycol) diacrylate (PEGDA), NPS, and Irgacure 819. This mixture created structures that have low non-specific adsorption and had good resolution in the z-axis. Additionally, the structures could be photocured with a broad-spectrum UV lamp after printing.
Despite high-resolution printers and materials, a new printing regime also had to be developed to achieve the desired channel width. Because of scattering, the area of crosslinked resin tended to be smaller than the actual pixel size. This meant that channels, which are essentially voids of non-exposed space, came out wider than designed. The solution that Gong et al. devised was to perform two print patterns for each exposed layer. The first print pattern exposes the entire area, save for the channel void and the second exposure only projects a pattern along the channel walls. This method enabled reproducible printing of channels with a ~20 µm width.
So how soon can you expect to get your hands on a 3D printer capable of printing true microchannels? While this report has been very promising, it is currently the only printer and only resin formulation with these capabilities. According to Greg Nordin, the corresponding author, the main challenge is to figure out “how to get this into people’s hands in a convenient way so they don’t have to mess around much to get it to work.” Like any new technology, for it to be truly useful, users want to spend more time making and less time fiddling. Fortunately, there already are markets for printing small, intricate parts, such as jewelry and dentistry, so it’s only a matter of time before manufacturers catch on to the market for 3D printed microfluidics.
To download the full article for free* click the link below:
Custom 3D printer and resin for 18 μm × 20 μm microfluidic flow channels
Hua Gong, Bryce P. Bickham, Adam T. Woolley and Gregory P. Nordin
Lab Chip, 2017
DOI: 10.1039/C7LC00644F
*Free to access until 29th August 2017.
About the Webwriter
Darius Rackus is a postdoctoral researcher at the University of Toronto working in the Wheeler Lab. His research interests are in combining sensors with digital microfluidics for healthcare applications.”