The human body consists of tens of trillions cells, all of which theoretically should have the same genome. Depending on genetic and environmental factors, some of these cells experience point mutations. Although most of those mutations are cleaned up by DNA repair enzymes, about 0.01% of them stay. A low percentage of the persistent mutations turn out to be ‘cancer’ while others stay recessive. Several genes, including the ones responsible for cell growth cycle, cause the persistent mutations. Uncontrolled growth of cells leads to formation of tumor, which is now prone to experience more mutations due to continuous proliferation. These mutations create heterogeneity among the cell population of a tumor, and eventually some cells leave their original tumor and start a new one in another organ of the body. When a cell leaves its parent tumor, it starts circulating in blood vessels before settling in. Those cells are called circulating tumor cells (CTCs), and around 1-10 CTC can be found in 1 mL of blood (which contains about 1 billion red blood cells, and 1 million white blood cells). Capturing ultra-rare CTCs has enormous implications in early cancer diagnosis. Often times, analysis of at least 10 mL of blood is necessary to capture sufficient CTCs to confirm their presence. The available technology can achieve CTC capturing in about 10 hours leading to the loss of target cells and decay of detection biomarkers.
Microfluidic devices are well known for precise sorting of microscale materials. Parallelizing microscale sorting in microfluidic devices enables high-throughput sample processing. In the light of this principle, David Issadore, Jina Ko, and their fellow researchers at University of Pennsylvania coined CaTCh FISH, a circulating tumor cell fluorescence in situ hybridization platform for rapid detection of CTCs. David Issadore kindly accepted to talk about this exciting lab-on-a-chip device. According to David, “the main strength of the CaTCh FISH is that it preserves the sensitivity and specificity of microfluidic cell sorting and RNA FISH, but through clever engineering allows these normally very slow laboratory-based operations to be performed rapidly and automatically on a chip.”
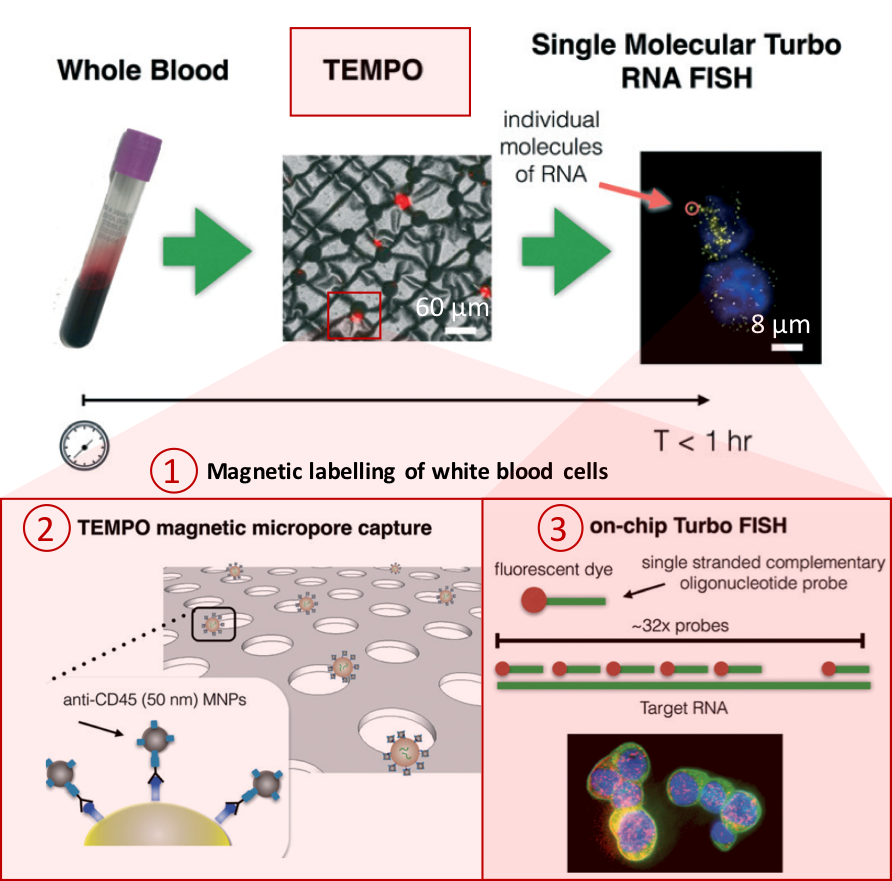
Figure 1. Overview of the CaTCh FISH platform. Whole blood sample is processed in TEMPO step, where magnetic nano particle based cell separation is followed by single-cell RNA analysis (modified from Ko et al., 2017).
Processing a whole blood sample using CaTCh FISH involves three steps (Figure 1). First, white blood cells are labelled with magnetic nanoparticles. Second, whole blood is passed through a magnetic micropore filter to selectively trap magnetically labelled cells. “Our magnetic micropore device rapidly and precisely removes all of the cells that we know are not CTCs”, says David. The operating principle of magnetic micropore filter is based on strong and highly localized microscale field gradients formed at the edge of micropores to enable application of high flow rates. Third, single cell RNA analysis is performed on isolated cells using rapid in-situ hybridization strategy so that CTCs can be identified within the isolated cell population. In this way, targeted CTCs can be isolated from the rest of the cell population regardless of their physical and molecular properties. Analysis of a 10 mL blood sample takes less than an hour.
As a key novelty, the researchers maintained high-throughput processing and high sensitivity at the same time by integrating the FISH technique (hybridization of 20-50 fluorescently-labelled oligonucleotide probes to the target RNA, and subsequent fluorescence-signal based detection to enhance signal-to-noise ratio) in a microfluidic chip. CaTCh FISH has also been tested in patients with pancreatic cancer and detected CTCs in the real patient samples.
“The CaTCH FISH technology can be easily modified to measure other rare cells, for the diagnosis of other cancers or for stem cell research for example, by modifying the RNA FISH probes”, says David. He considers converting the platform to a high-precision hospital-based diagnostic tool and he collaborates with a company in the Bay area for this process. The CaTCh FISH device is poised to have a big impact on the way cancer is diagnosed.
To download the full article for free* click the link below:
Jina Ko, Neha Bhagwat, Stephanie S. Yee, Taylor Black, Colleen Redlinger, Janae Romeo, Mark O’Hara, Arjun Raj, Erica L. Carpenter, Ben Z. Stanger and David Issadore
Lab Chip, 2017, Paper
DOI: 10.1039/C7LC00703E
Burcu Gumuscu is a postdoctoral fellow in Herr Lab at UC Berkeley in the United States. Her research interests include development of microfluidic devices for quantitative analysis of proteins from single-cells, next generation sequencing, compartmentalized organ-on-chip studies, and desalination of water on the microscale.
*until 5th January 2018











