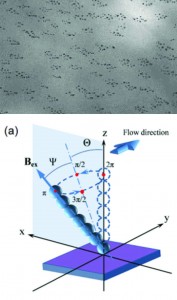
Cilia are microscale ‘eyelash-like’ extensions of eukaryotic cells found in epithelial linings throughout the body. In the fallopian tubes, windpipe, and lungs, motile cilia beat rhythmically to move objects within the viscous liquid above. Non-motile cilia in the inner ear transduce mechanical vibrations to electrical signals to ultimately excite auditory nerves.
Previously, these appendages have been built using advanced microfabrication techniques. Now, for the first time, researchers at Eindhoven University of Technology in the Netherlands present a simple bench-top fabrication method for self-assembly of artificial cilia using magnetic beads and latex particles.
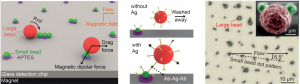
Jaap den Toonder and his team created the artificial cilia sans cleanroom by coating magnetic beads with latex particles in a fluid cell using a magnetic field to control bead orientation. Latex particles were attracted to the beads by electrostatic forces and the whole structure was bonded together using a heating cycle. The completed artificial cilia were 3 μm in diameter and could be made into lengths of up to 33 μm by optimizing the magnetic field strength and protocol duration. The cilia were actuated by oscillating magnetic fields after fabrication to produce flow velocities of 3 μm/s.
Microfluidic devices operate under low Reynolds numbers where inertia is negligible, presenting a significant challenge to efficient mixing and moving of objects. Cilia and flagella evolved in organisms living in low Reynolds numbers to enable swimming by generating fluid flow using nonreciprocal (nonreversible) motions of beating and twisting.1, 2 The fabrication method presented in this work powerfully enables artificial cilia to be fabricated in situ in assembled platforms and “ship-in-a-bottle” constructed devices, thus facilitating practical applications for these structures in existing microfluidic platforms for bio-inspired fluid manipulation at the microscale.
Out of the cleanroom, self-assembled magnetic artificial cilia, Ye Wang, Yang Gao, Hans Wyss, Patrick Anderson, and Japp den Toonder, Lab Chip, 2013, 13, 3360-3366. DOI: 10.1039/C3LC50458A
References:
1. E. M. Purcell, AIP Conference Proceedings, 1976, 28, 49.
2. S. Khaderi, J. den Toonder and P. Onck, Biomicrofluidics, 2012, 6, 014106.