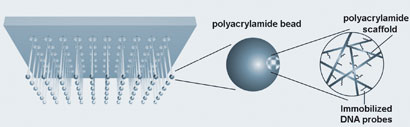
A microfluidic chip that can come up with a DNA profile in less than three hours has been designed by US scientists for use at crime scenes.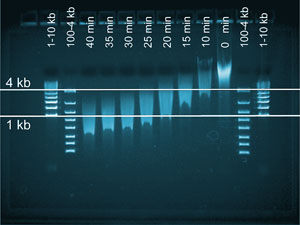
With current techniques, forensic scientists have to wait up to eight hours to get results. Using microchips to speed up the process has been investigated but integrating all of the profiling steps in one device has remained elusive until now. Richard Mathies from the University of California, Berkeley, and colleagues, in collaboration with the US Department of Justice, have produced a portable method to test DNA at a crime scene that integrates all of the steps in one device.
Andy Hopwood, an expert in DNA analysis techniques from the UK’s Forensic Science Service, believes that the work is ‘without a doubt a very exciting and significant development toward the total integration of the DNA-based human identification process onto a single microchip’.
Read Holly Sheahan’s Chemistry World article online here or go straight to the HOT Lab on a Chip paper:
Integrated DNA purification, PCR, sample cleanup, and capillary electrophoresis microchip for forensic human identification
Peng Liu, Xiujun Li, Susan A. Greenspoon, James R. Scherer and Richard A. Mathies
Lab Chip, 2011, 11, 1041
DOI: 10.1039/c0lc00533a