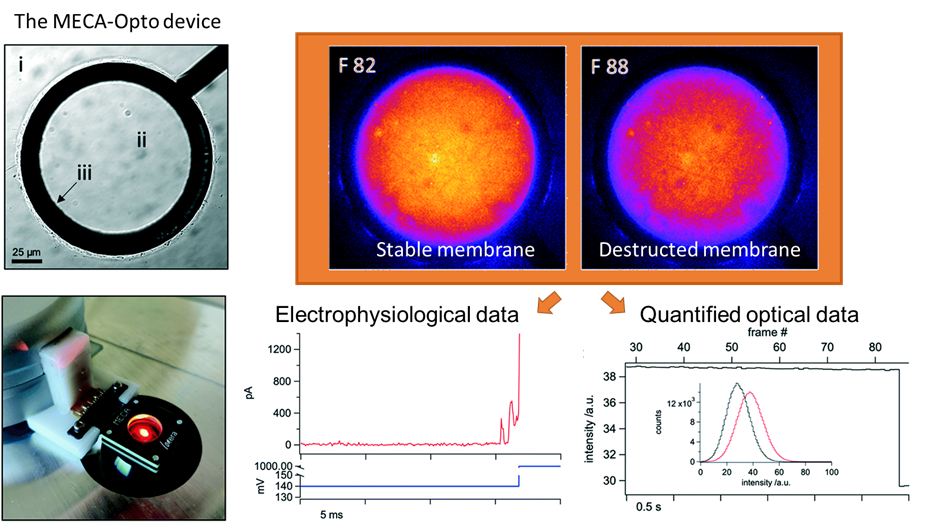
We are delighted to welcome:
- Professor Xingyu Jiang (SUSTech, China)
- Professor Amy Herr (UC Berkeley, USA)
- Professor Hongkai Wu (HKUST, Hong Kong)
as our newest Lab on a Chip Associate Editors! You can read more about them and their research focus below.

A my Herr
my Herr
John D. & Catherine T. MacArthur Professor, Department of Bioengineering,
University of California, Berkeley, USA
Amy Herr is the John D. & Catherine T. MacArthur Professor in the Department of Bioengineering , a Chan Zuckerberg (CZ) Biohub Investigator, and CTO of the Chan Zuckerberg Biohub Network.
A major focus of the Herr lab is engineering innovation for analysis of complex biological systems — as is required to address questions important to both fundamental biological systems and applied clinical research. They employ a combination of approaches drawn from chemical engineering, mechanical engineering, and electrical engineering with strong foundations in biology, materials science, and analytical chemistry. In essence, they strive to advance the “mathematization” of biology & medicine. Their research projects span understanding fundamental transport to materials design to applications in life sciences tools and diagnostics.
Professor Herr’s research has been recognized by prominent international andnational organizations, including: 2018 Sciex Microscale Separations Innovation Medal, 2017 Georgina Sweet Lectureship from the Australian Research Council, 2016 Mid-career Achievement Award from the American Electrophoresis Society, 2015 Georges Guiochon Faculty Fellow from HPLC, 2012 Young Innovator Award from Analytical Chemistry/CBMS, 2011 NSF CAREER award, 2010 NIH New Innovator Award, 2010 Alfred P. Sloan Research Fellowship in chemistry, 2010 New Investigator Award in Analytical Chemistry from Eli Lilly & Co., and a 2009 Defense Advanced Research Projects Agency (DARPA) Young Faculty Award. Her commitment to creating a strong professional community is reflected in recognition as: 2019 Award for Excellence in Postdoctoral Mentoring, Visiting Scientist & Posdoc. Assoc. at UC Berkeley, 2017 Berkeley Visionary Award from the City of Berkeley Chamber of Commerce, and 2007 Outstanding Mentor Award from Sandia National Labs.
Research Expertise and Interest
microfluidics, bioanalytical separations, diagnostics, electrokinetic transport, engineering design
Find out more on the Herr Lab website – Bioinstrumentation for Quantitative Biology & Medicine
Here are a selection of Amy’s most recent Lab on a Chip papers below:
Xingyu Jiang
 Chair Professor, Department of Biomedical Engineering Southern University of Science and Technology, China
Chair Professor, Department of Biomedical Engineering Southern University of Science and Technology, China
Xingyu Jiang is a Chair Professor at the Southern University of Science and Technology, Shenzhen, China. He obtained his BS at the University of Chicago (1999) and PhD at Harvard University (Chemistry, 2004). In 2005, he joined the National Center for NanoScience and Technology/the University of the Chinese Academy of Sciences. He moved to the Southern University of Science and Technology in 2018. His research interests include microfluidics and nanomedicine and their applications in diagnostics, screening for therapeutics, as well as engineered tissues. He has over 300 publications in peer-reviewed journals. He was awarded the “Hundred Talents Plan” of the Chinese Academy of Sciences, the National Science Foundation of China’s Distinguished Young Scholars Award, the Scopus Young Researcher Gold Award, and the Human Frontier Science Program Young Investigator Award. He is a Fellow of the Royal Society of Chemistry (UK) and the American Institute of Medical and Biological Engineering.
Xingyu has been a committed member of the Lab on a Chip Editorial Board since 2020, and will now start handling papers for the journal.
Research Expertise and Interest
microfluidics & nanomedicine, and their applications in diagnostics, screening for therapeutics, engineered tissues
Find out more on the Jiang lab website and follow @xingyu on Twitter
Here are a selection of Xingyu’s most recent Lab on a Chip papers below:
 Hongkai Wu
Hongkai Wu
Professor, Department of Chemistry, Department of Chemical & Biological Engineering
Hong Kong University of Science and Technology, Hong Kong
Hongkai Wu is a Professor in Department of Chemistry & Department of Chemical & Biological Engineering at the Hong Kong University of Science and Technology, Hong Kong, China. Dr. Wu received his BSc from University of Science and Technology of China in 1995 and Ph.D from Harvard University, USA in 2002. After working as a Postdoctoral Fellow at Stanford, he joined the faculty in Department of Chemistry in Tsinghua University, Beijing, China in 2005 and moved to HKUST as an Assistant Professor in 2007. In 2012, he was promoted to be an Associate Professor. From 2009 to 2015, he established a satellite laboratory in Tohoku University, Sendai, Japan, focusing on developing functional biomaterials.
Research at the Wu lab focuses on the interdisciplinary frontiers of microfluidics, bioanalytical science and materials chemistry. They use the technologies in MEMS, microfluidics, soft lithography, and surface chemistry to design and provide new tools for the applications and understanding of fundamentals in materials and biological sciences, including microfluidic chemical reactors, high throughput single-cell analysis and chemical separations.
Research Expertise and Interest
microfluidics, bioanalytical science, materials chemistry, single-cell analysis
Find out more on the Wu lab website
Here are a selection of Hongkai’s Lab on a Chip papers below:
Please join us in welcoming our new Associate Editors. We look forward to continuing to work with them on Lab on a Chip, and they are looking forward to handling your papers!
Submit your paper today!
Comments Off on Welcome to our newest Lab on a Chip Associate Editors in 2023!























 This Lectureship honours and supports the up and coming, next generation of scientists who have significantly contributed to the understanding or development of miniaturised systems.
This Lectureship honours and supports the up and coming, next generation of scientists who have significantly contributed to the understanding or development of miniaturised systems.