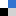Symbiotic Actinomycetes are known to produce a plethora of natural products of pharmacological importance and global genome sequencing efforts unraveled their unique and still mostly untapped biosynthetic potential. To activate natural product biosynthesis in Actinobacteria and to harvest the wealth of natural products, cultivation conditions mimicking the natural environment turned out to be amongst the most promising approach next to molecular biotechnological tools. We have recently applied ecology-driven interaction studies between bacterial symbionts of fungus-farming termites and their natural antagonists to activate the biosynthetic repertoire of actinomycetous symbionts. In light of these studies, we revisited the bacterial symbiont Streptomyces sp. M56 (from now on named M56), which showed very strong antagonistic effect against parasitic co-isolated fungi and was found to produce, not only the ansamycin-derived natural products natalamycin and geldanamycin, but also elaiophylins and the structurally related efomycins.
Our group thoroughly investigated culture extracts obtained from strain M56 by liquid chromatography/high resolution tandem mass spectrometry (LC-HRMS2) and Global Natural Product Social Molecular Networking (GNPS) analysis. Indeed, dereplication of the acquired MS2 data based on available databases resulted in the detection of several polyhydroxylated macrolides identified as the antifungal azalomycins (Figure 1). Although the planar structures of azalomycins were reported as early as 1959 and the first skeletal structure assigned already in 1982, reports on their absolute structures remained inconclusive with varying structural assignments across the literature (Figure 2).
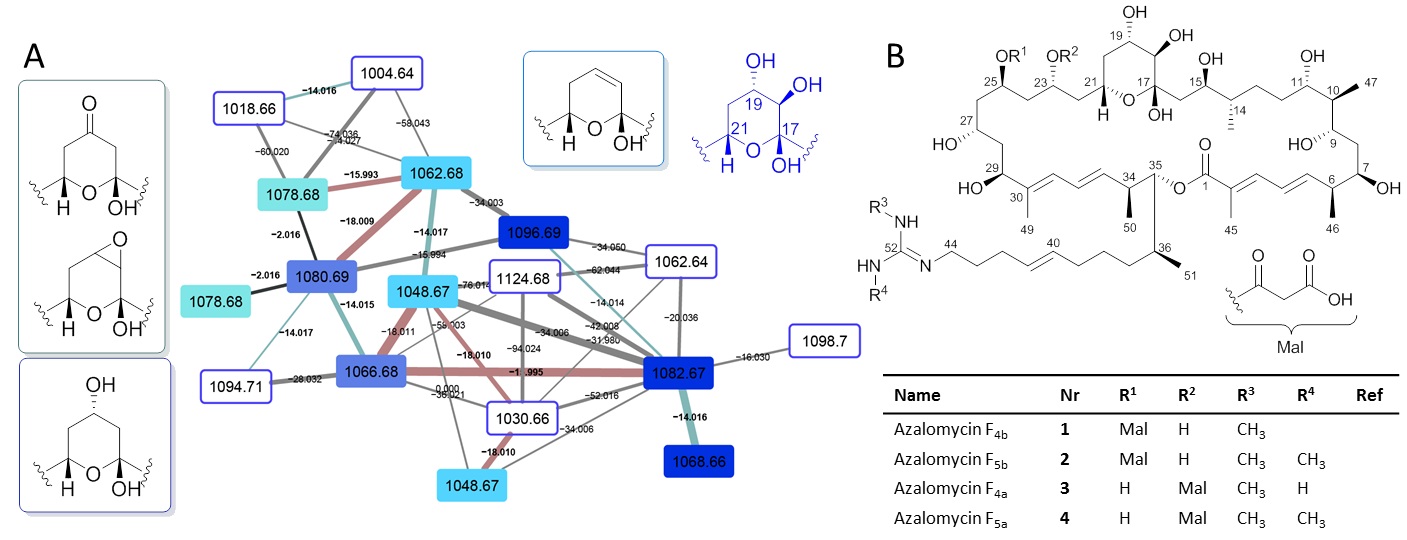
Figure. 1. A) HRMS2-based GNPS analysis depicting molecular ion cluster putatively assigned as an azalomycin cluster with putative structural features assigned to m/z 1096.69, 1082.67 and 1068.66 (diol moiety C-18 and C-19); m/z 1080.69 and 1066.68 (C-19 alcohol); m/z 1062.68 and 1048.67 (enoyl derivatives); m/z 1078.68 (C-19/C-18 epoxy). Data obtained from HRMS2 measurements of EtOAc extract (7 d, ISP2 liquid broth) in positive mode ESI-HRMS. B) Stereochemical assignment of isolated azalomycins (1–4) from Streptomyces sp. M56.
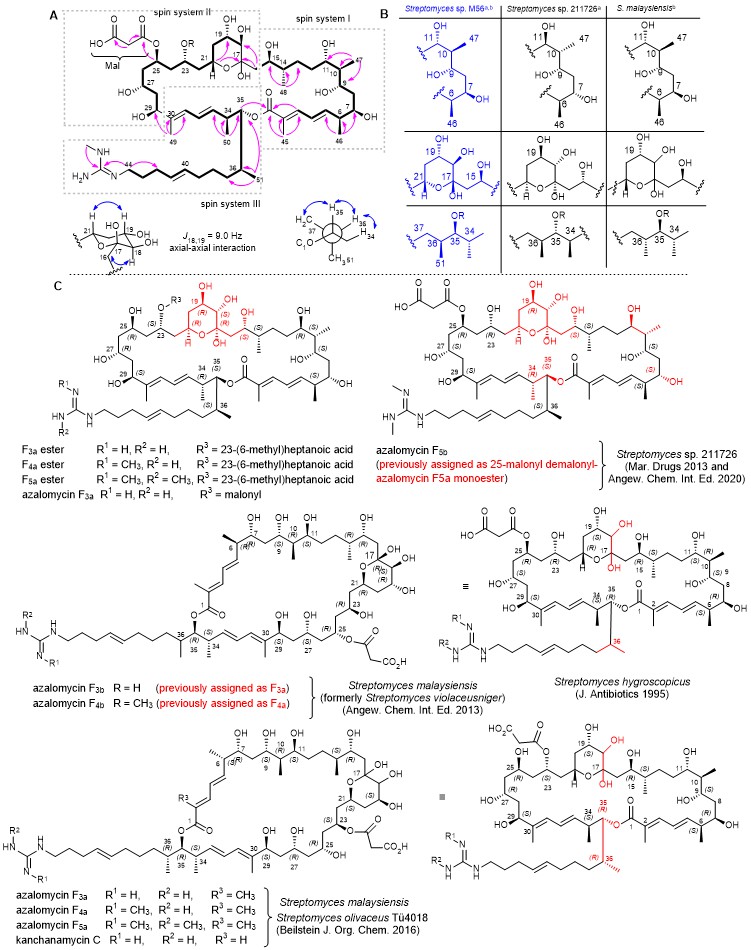
Figure 2. A) Structure of azalomycin (1) with key COSY (bold) and HMBC (pink arrow) and NOE (blue double-headed arrow) correlations and coupling constant analysis. B) Stereochemical assignment of azalomycins substructures isolated from different producer strains based on a) NMR analysis and b) in silico PKS domain predictions. C) Chemical structures of azalomycins reported from Streptomyces sp. 211726,22,29 and Streptomyces malaysiensis DSM 413723 and Streptomyces hygroscopicus. Stereocenters that differ from herein proposed structure or have been newly assigned based on analytical experiments are colour-coded in red.
Due to their strong antimicrobial activity, but inconclusive structural assignments, we re-evaluated the analytical and bioactivity data of azalomycins. Here, we provide a conclusive analysis of their stereochemical assignment based on comparative NMR and gene clusters studies and propose to partially revise the absolute structures and names of azalomycins from previous reports (Figure 3).
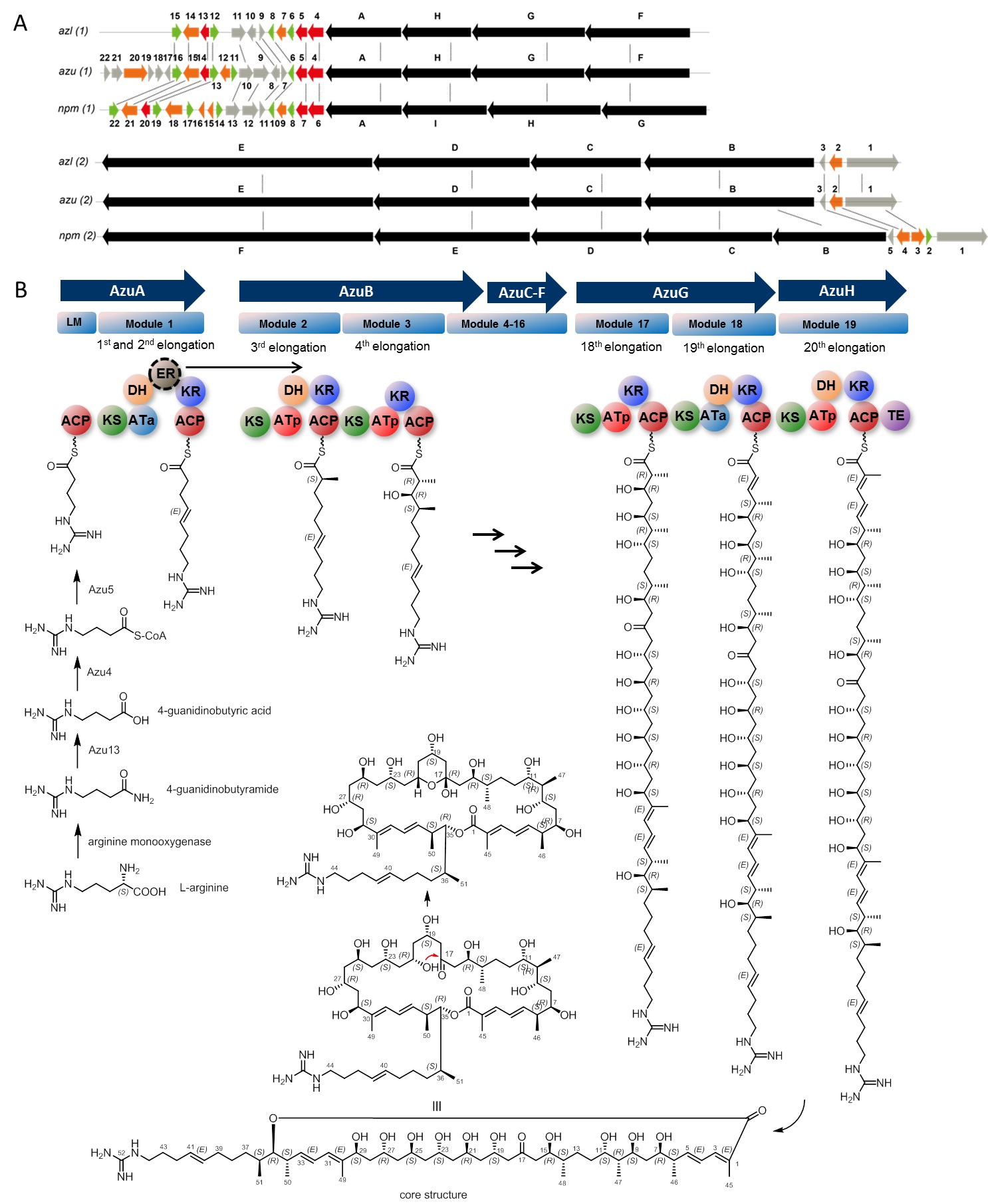
Figure 3. A) Graphical comparison of azalomycins biosynthesis gene cluster (azu) from Streptomyces sp. M56 and azalomycin F3a biosynthesis gene cluster (azl) from Streptomyces sp. 211726, and niphimycin C biosynthesis gene cluster (npm) from Streptomyces sp. IMB7-145. B) Schematic representation of important biosynthetic transformations yielding the azalomycin core structure. The ER domain of module 1 is assumed to be non-functional; dash-circulated domain represents the missing module required for chain elongation. Ata (blue): malonyl-CoA selective AT domain (acetate unit); ATp (light red): methylmalonyl-CoA selective AT domain (propionate unit); ACP (dark red): acyl carrier protein; DH (light orange): dehydratase; ER (brown-black): enoylreductase; KR: ketoreductase (A1 or B1); TE (purple): thioesterase.
Ki Hyun Kim is associate professor in the School of Pharmacy at the Sungkyunkwan University (Republic of Korea). He earned his pharmacist’s license in 2005 and he received his PhD in the School of Pharmacy at the Sungkyunkwan University in 2011. From 2011 to 2012 he did postdoctoral studies at the School of Pharmacy at the Sungkyunkwan University, and from 2012 to 2013 at Harvard University and Harvard Medical School. He started his faculty in the School of Pharmacy at the Sungkyunkwan University in 2014. The main research field of Professor Kim is the discovery of the secondary metabolites from diverse natural resources including wild mushrooms, insect-associated bacteria, marine natural sources and medicinal plants and his group has substantial experience and expertise in natural product isolation and structural elucidation using mainly NMR-based analysis as well as biological activity evaluations. He is the author of 7 patents and more than 300 articles indexed by SCIE and cited more than 1500 times with an index H = 20.