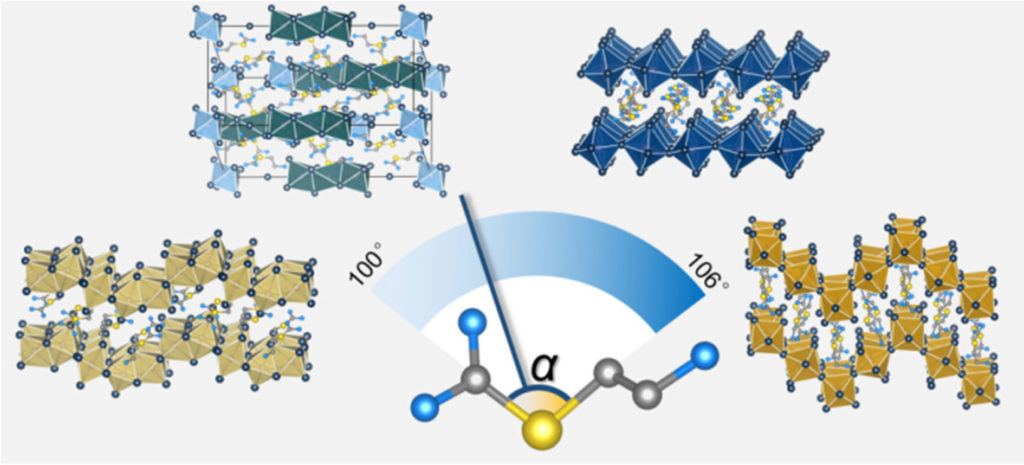
Zirconium-89 complexation chemistry is an important area of research in the context of developing radiolabelled proteins for applications in diagnostic positron emission tomography (PET) imaging. For this imaging technology, the metalloradionuclide 89Zr4+ ion needs to be sequestered by a ligand to form a coordination complex that is thermodynamically, kinetically, and metabolically stable in biological systems. In this regard, desferrioxamine B (DFO), a natural bacterial siderophore, is one of the outstanding hexadentate linear chelator for zirconium-89, used in clinical trials with 89ZrDFO-radiolabeled antibodies (mAbs). Nevertheless, preclinical studies have demonstrated that 89ZrDFO-mAbs can suffer from dissociation and metal ion release in vivo resulting in partial bone uptake in mice which could be partially due to the incomplete coordination sphere around the metallic cation. Driven by the goal of increasing the stability of the 89Zr4+ coordination complex toward demetallation in vivo, several groups around the world have explored the synthesis and coordination chemistry of novel multidentate chelates with coordination numbers from 6 to 8 but the development of heptadentate remained unexplored.
Recently, a collaborative work between the group of Prof. Dr Jason P. Holland (University of Zurich, Switzerland) and a team from the Institut Plurisdisciplinaire Hubert Curien (IPHC, CNRS, University of Strasbourg, France) have demonstrated that photoactivatable heptadentate chelates could be a new alternative for the ultra-fast, light-induced production of stable 89Zr-mAbs in vivo (Figure 1). The researchers synthesise new chelates, used density functional theory to predict the thermodynamic stability, and studied the in vitro stability of the radiolabelled complexes to find the most promising candidate for in vivo application.
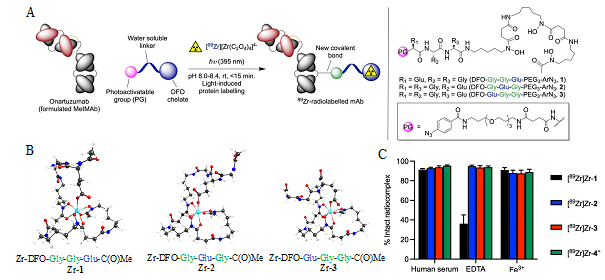
Figure 1. (A) Overview of the light-induced photoradiosynthesis to produce 89Zr-labelled monoclonal antibodies (mAbs) and structure of the ligands (1 – 3). (B) Optimised structures of the three model Zr complexes. (C) Bar chart showing the stability of the 89Zr-radiolabelled complexes (formed from chelates 1 – 4) under different challenge conditions.
The researchers also selected the most stable complex (Zr-2) and produced 89Zr-radiolabelled onartuzumab (the monoclonal antibody component of MetMAbTM which binds to the human hepatocyte growth-factor receptor c-MET) using photoradiochemical methods. Finally, the pharmacokinetic profile and c-MET targeting was evaluated in vivo and ex vivo by using PET imaging and biodistribution studies in female athymic nude mice bearing subcutaneous MKN-45 human gastric cancer xenografts (Figure 2).
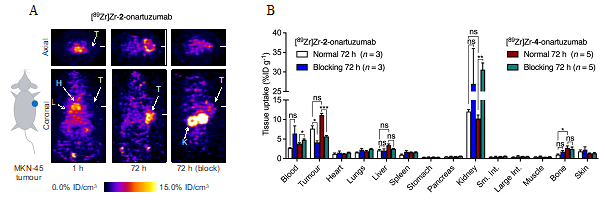
Figure 2. (A) Coronal and axial PET images taken through the centre of the tumours showing the spatial distribution of [89Zr]Zr-2-onartuzumab over time after intravenous administration in mice bearing subcutaneous MKN-45 tumours on the right flank. T = Tumour, H = Heart, L = Liver, K = Kidneys. (B) Bar chart showing ex vivo biodistribution data (%ID g-1) for the uptake of [89Zr]Zr-2-onartuzumab (normal group, white; blocking group, blue) and the 6-coordinate control compound [89Zr]Zr-4-onartuzumab (normal group, red; blocking group, green) in mice bearing MKN-45 tumours.
Overall, the researchers proved that [
89Zr]Zr-
2-onartuzumab provides specific tumour targeting and high tumour-to-organ contrast on the PET pictures and from the biodisitribution analysis. The results obtained in the study confirm that heptadentate complexes of
89Zr display improved stability
in vivo compared with hexadentate analogs and are promising candidates for future
89Zr-radiotracer design.
About the corresponding author

Jason P. Holland is from Yorkshire in the UK and is currently an SNSF Professor for Medicinal Radiochemistry at the University of Zurich. Research activities in the Holland group focus on advancing radiolabelling methods through novel bioconjugation approaches for labelling bioactive molecules with various radionuclides (18F, 64Cu, 67/68Ga, 86/90Y, 99mTc, 111In, 177Lu, 188Re, etc).
E-mail: jason.holland@chem.uzh; Twitter: @HollandLab_