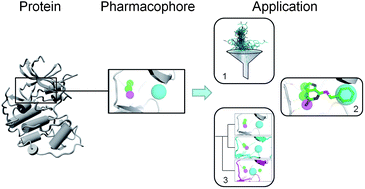
The review covers (i) protein structure preparation, (ii) binding site detection, (iii) pharmacophore feature definition, and (iv) pharmacophore feature selection, concluding that ‘SBPs are valuable tools for hit and lead optimization, compound library design and target hopping, especially in cases where ligand information is scarce‘.
From the protein’s perspective: the benefits and challenges of protein structure-based pharmacophore modeling
Marijn P. A. Sanders, Ross McGuire, Luc Roumen, Iwan J. P. de Esch, Jacob de Vlieg, Jan P. G. Klomp and Chris de Graaf
DOI: 10.1039/C1MD00210D