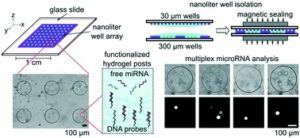
MIT researchers develop a simple new chip that simplifies sample preparation and reduces sample volume for micro-RNA analysis
In recent years, microRNAs (miRNAs) have emerged as potential biomarkers for a range of diseases. These short (~22 nucleotide) sequences of non-coding RNA function by silencing messenger RNA and thus, provide a form of post-transcriptional regulation in the cell. Consequently, their regulation can have a significant impact on cell function. While they are implicated in many diseases, their role in cancer is of particular interest. It is known that multiple miRNAs are dysregulated in tumours compared to healthy tissues and they can be used as biomarkers in cancer diagnosis. Testing for multiple miRNAs provides a better diagnosis, so it is important to be able to test them as a panel. There are many techniques available to test for miRNAs such as gene chips, quantitative RT-PCR, and RNA-seq, but these technologies all have their own shortcomings. What is common to all them is the need for sample preparation, and mainly RNA extraction from the tissue or cells to be tested. In their recent HOT article, Tentori and researchers from the Doyle Group at MIT describe a chip that could overcome this problem and perform multiplexed testing of miRNA all within a minimum volume of just a few nanoliters.
The new chip comprises two glass slides which can be sandwiched together. The bottom slide has an array of 300 µm diameter wells which contain the miRNA sensors and serve to isolate samples into a small reaction volume. The top slide is then used to deliver lysis reagents to the sample. The authors tried a variety of designs for the top plate, but ultimately settled on an array of 30 µm diameter wells. This has two advantages; 1) Reagents are precisely metered and 2) the top and bottom plates can be sandwiched robustly without any need for precision handling or alignment. This last point is really important to Augusto Tentori, a postdoc in the Doyle Group and the lead author of the paper. Tentori wants “to make devices that are simple and robust, so translation is easier.” The miRNA sensors are polyethylene glycol diacrylate (PEGDA) hydrogels that contain complementary DNA probes. The posts are photopolymerized in the wells and various sizes, shapes, and patterns of posts can be made. Further, a single well can contain a variety of posts, each functionalized with a different DNA probe targeting a different miRNA. In this way, multiplexed assays can be performed with spatial separation. The chip format of the assay is more sensitive than previous formats and can detect miRNA from less than 20 cells.
Tentori is really excited about the prospects of this new chip. His co-authors include pathologists who have been guiding the project to make sure it is clinically useful, and he really wants to see this technology get into the hands of pathologists and diagnostic technicians.
Read the full article by Tentori et al. here “Quantitative and multiplex microRNA assays from unprocessed cells in isolated nanoliter well arrays” that features in Lab on a Chip’s HOT article collection.
Darius Rackus (right) is a postdoctoral researcher in the Dittrich Bionalytics Group at ETH Zürich. His research interests are in developing integrated microfluidic tools for healthcare and bioanalysis