These HOT articles were recommended by our referees and are free to access for 4 weeks*
Antimicrobial susceptibility assays in paper-based portable culture devices
Frédérique Deiss, Maribel E. Funes-Huacca, Jasmin Bal, Katrina F. Tjhung and Ratmir Derda
Lab Chip, 2014,14, 167-171
DOI: 10.1039/C3LC50887K, Communication
Cell force measurements in 3D microfabricated environments based on compliant cantilevers
Mattia Marelli, Neha Gadhari, Giovanni Boero, Matthias Chiquet and Jürgen Brugger
Lab Chip, 2014,14, 286-293
DOI: 10.1039/C3LC51021B, Paper
A differential dielectric affinity glucose sensor
Xian Huang, Charles Leduc, Yann Ravussin, Siqi Li, Erin Davis, Bing Song, Dachao Li, Kexin Xu, Domenico Accili, Qian Wang, Rudolph Leibel and Qiao Lin
Lab Chip, 2014,14, 294-301
DOI: 10.1039/C3LC51026C, Paper
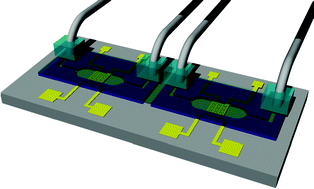
Microfluidic transwell inserts for generation of tissue culture-friendly gradients in well plates
Christopher G. Sip, Nirveek Bhattacharjee and Albert Folch
Lab Chip, 2014,14, 302-314
DOI: 10.1039/C3LC51052B, Paper
From themed collection Lab on a Chip Top 10%
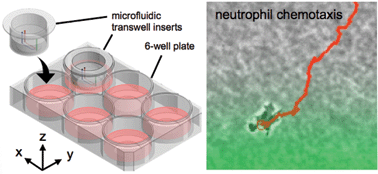
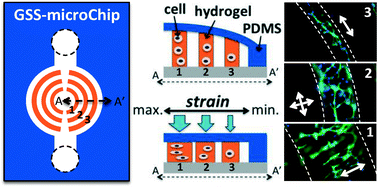
Gradient static-strain stimulation in a microfluidic chip for 3D cellular alignment
Hsin-Yi Hsieh, Gulden Camci-Unal, Tsu-Wei Huang, Ronglih Liao, Tsung-Ju Chen, Arghya Paul, Fan-Gang Tseng and Ali Khademhosseini
Lab Chip, 2014,14, 482-493
DOI: 10.1039/C3LC50884F, Paper
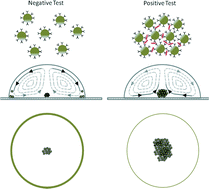
Biosensor design based on Marangoni flow in an evaporating drop
Joshua R. Trantum, Mark L. Baglia, Zachary E. Eagleton, Raymond L. Mernaugh and Frederick R. Haselton
Lab Chip, 2014,14, 315-324
DOI: 10.1039/C3LC50991E, Paper
From themed collection Lab on a Chip Top 10%
Flow of suspensions of carbon nanotubes carrying phase change materials through microchannels and heat transfer enhancement
Sumit Sinha-Ray, Suman Sinha-Ray, Hari Sriram and Alexander L. Yarin
Lab Chip, 2014,14, 494-508
DOI: 10.1039/C3LC50949D, Paper
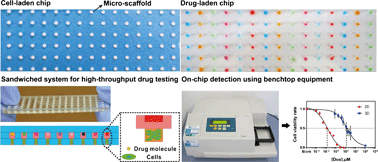
Micro-scaffold array chip for upgrading cell-based high-throughput drug testing to 3D using benchtop equipment
Xiaokang Li, Xinyong Zhang, Shan Zhao, Jingyu Wang, Gang Liu and Yanan Du
Lab Chip, 2014,14, 471-481
DOI: 10.1039/C3LC51103K, Paper
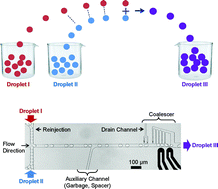
Synchronized reinjection and coalescence of droplets in microfluidics
Manhee Lee, Jesse W. Collins, Donald M. Aubrecht, Ralph A. Sperling, Laura Solomon, Jong-Wook Ha, Gi-Ra Yi, David A. Weitz and Vinothan N. Manoharan
Lab Chip, 2014,14, 509-513
DOI: 10.1039/C3LC51214B, Paper
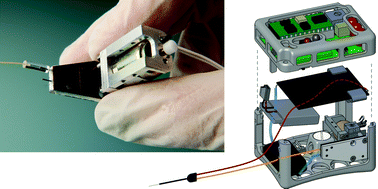
A microfluidic reciprocating intracochlear drug delivery system with reservoir and active dose control
Ernest S. Kim, Erich Gustenhoven, Mark J. Mescher, Erin E. Leary Pararas, Kim A. Smith, Abigail J. Spencer, Vishal Tandon, Jeffrey T. Borenstein and Jason Fiering
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51105G, Paper
From themed collection Lab on a Chip Top 10%
Impedance matched channel walls in acoustofluidic systems
Ivo Leibacher, Sebastian Schatzer and Jürg Dual
Lab Chip, 2014,14, 463-470
DOI: 10.1039/C3LC51109J, Paper

Magnetoactive sponges for dynamic control of microfluidic flow patterns in microphysiological systems
Sungmin Hong, Youngmee Jung, Ringo Yen, Hon Fai Chan, Kam W. Leong, George A. Truskey and Xuanhe Zhao
Lab Chip, 2014,14, 514-521
DOI: 10.1039/C3LC51076J, Paper
From themed collection Lab on a Chip Top 10%

The microfluidic post-array device: high throughput production of single emulsion drops
E. Amstad, S. S. Datta and D. A. Weitz
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51213D, Paper
From themed collection Lab on a Chip Top 10%
Interdroplet bilayer arrays in millifluidic droplet traps from 3D-printed moulds
Philip H. King, Gareth Jones, Hywel Morgan, Maurits R. R. de Planque and Klaus-Peter Zauner
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51072G, Paper

A 1024-sample serum analyzer chip for cancer diagnostics
Jose L. Garcia-Cordero and Sebastian J. Maerkl
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51153G, Paper
From themed collection Lab on a Chip Top 10%
Utilization and control of bioactuators across multiple length scales
Vincent Chan, H. Harry Asada and Rashid Bashir
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC50989C, Critical Review
Microfabricated perfusable cardiac biowire: a platform that mimics native cardiac bundle
Yun Xiao, Boyang Zhang, Haijiao Liu, Jason W. Miklas, Mark Gagliardi, Aric Pahnke, Nimalan Thavandiran, Yu Sun, Craig Simmons, Gordon Keller and Milica Radisic
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51123E, Paper
From themed collection Lab on a Chip Top 10%
Radiolabelling diverse positron emission tomography (PET) tracers using a single digital microfluidic reactor chip
Supin Chen, Muhammad Rashed Javed, Hee-Kwon Kim, Jack Lei, Mark Lazari, Gaurav J. Shah, R. Michael van Dam, Pei-Yuin Keng and Chang-Jin “CJ” Kim
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51195B, Paper
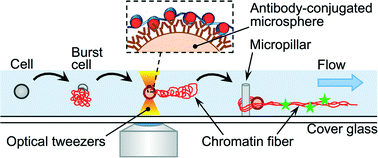
Non-destructive handling of individual chromatin fibers isolated from single cells in a microfluidic device utilizing an optically driven microtool
Hidehiro Oana, Kaori Nishikawa, Hirotada Matsuhara, Ayumu Yamamoto, Takaharu G. Yamamoto, Tokuko Haraguchi, Yasushi Hiraoka and Masao Washizu
Lab Chip, 2014, Advance Article
DOI: 10.1039/C3LC51111A, Paper
*Free access to individuals is provided through an RSC Publishing personal account. It’s quick, easy and more importantly – free – to register!