Mapping the human genome project has been one of the world’s largest scientific collaborations. Completing the full genome sequencing for “the book of life” took more than 10 years with the efforts of 1000’s scientists and a budget of $3 billion. About 20 years after the finalization of this enormous project, it is now possible to complete a full human genome sequencing within 8 days for about $1,000 thanks to more advanced sequencing tools. Further improvements in genome sequencing tools are still warranted today because the genome sequencing field has been embraced by many more applications, including forensics, disease modeling & identification, and personalized medicine (e.g., identifying the genes that cause a medicine to work in one patient but not in another).
Initial sequencing technologies relied on standard DNA electrophoresis techniques such as slab gels and capillaries, allowing for the preparation of only small numbers of samples at a time. The sample preparation limitation was the primary reason for the increased costs and processing duration during the human genome project. Many efforts have been directed towards improving sample preparation techniques in the last decades. As the first step, electrophoresis techniques have been optimized to boost the sample throughput with user-friendly, smaller, and functional platforms. Traditional DNA separation gels, which have been used as the golden standard for many decades, have been replaced by microfabricated post arrays and nanometer-scale deterministic lateral displacement arrays.
A nice example of nanometer-scale deterministic lateral displacement arrays has been demonstrated recently by researchers from IBM T.J. Watson Research Center and Icahn School of Medicine at Mount Sinai in New York, USA. In this work, the researchers fractioned DNA in the range of 100-10.000 base pairs with a size-selective resolution of 200 base pairs. To achieve that, four different microchip configurations were fabricated on silicon wafers, where the array and nanopillar sizes were tuned for each configuration to obtain the optimum separation performance for the selected range of DNA fragments. Each configuration contained several separate arrays to conduct independent runs in a single chip. Instead of applying an electric field, the researchers applied pressure-driven force to separate the fragments. This strategy is particularly useful for separating non-charged species without being affected by buffer conditions (e.g., ionic strength).
| The separation mechanism in the nanometer-scale deterministic lateral displacement array is simple: If the size of a DNA fragment is larger than the diameter of the pillars, the fragment is deflected towards the collection wall at a large angle, also called the bump mode. If the size of a DNA fragment is smaller than the diameter of the pillars, the fragment migrates at an angle nominally zero, termed as the zig zag mode. In such a system, diffusion of DNA fragments lead to intermediate migration angles, termed as the partial-bump mode. Different sizes of DNA fragments could be separated in the array since the fragments will follow distinct trajectories thanks to the existence of different modes. Figure 1 summarizes the separation mechanisms and gives an outline for the nanometer-scale deterministic lateral displacement array. |
In the nanometer-scale deterministic lateral displacement array, the gap sizes were tuned from microscale to nanoscale only, without application of any other molecules that could change the DNA diffusion behavior, ionic strength (changing the effective gap distances). In such a setting, the researchers identified, for the first time, the flow velocity-dependence of different fragment lengths. Mainly, changing flow velocity caused a transition between bump and zig zag modes for the given size range of different DNA fragments: Slow speeds lead to partial-bump mode, and high speeds lead to the collapse of all DNA fragments to zigzag mode. The nanometer-scale deterministic lateral displacement array could also be used as a purification tool with 75% recovery and 3-fold concentration enhancement of DNA fragments. This tool could be used effectively for preparing next-generation sequencing libraries, on-chip DNA characterization, and circulating DNA characterization applications.
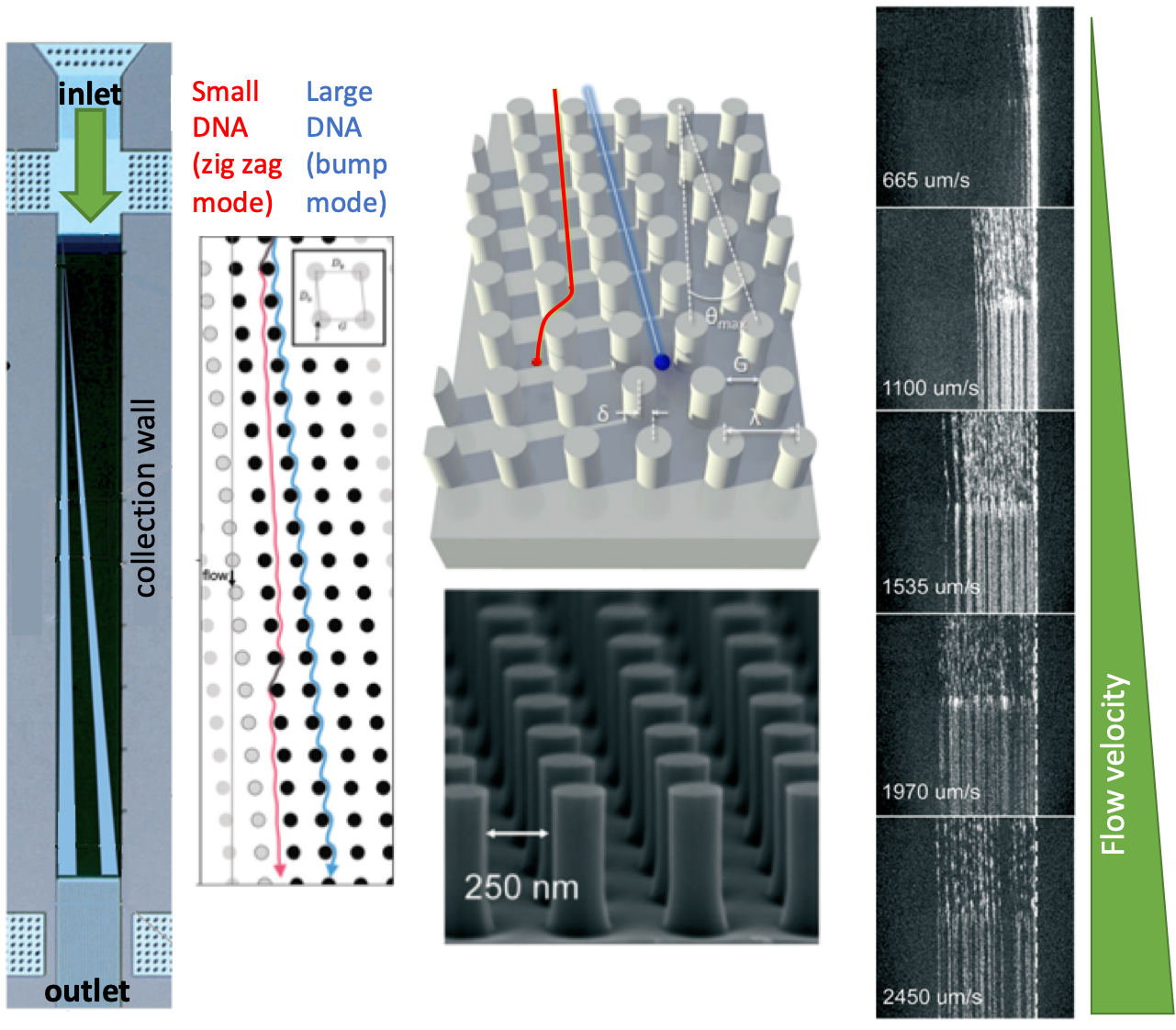
Figure 1. The nanometer-scale deterministic lateral displacement array and DNA separation mechanism at different flow velocities.
To download the full article for free* click the link below:
Gel-on-a-chip: continuous, velocity-dependent DNA separation using nanoscale lateral displacement
Benjamin H. Wunsch, Sung-Cheol Kim, Stacey M. Gifford, Yann Astier, Chao Wang, Robert L. Bruce, Jyotica V. Patel, Elizabeth A. Duch, Simon Dawes, Gustavo Stolovitzky and Joshua T. Smith, Lab Chip, 2018, Lab on a Chip Articles
DOI: 10.1039/c8lc01053f
Burcu Gumuscu is a researcher in Mesoscale Chemical Systems Group at the University of Twente in the Netherlands. Her research interests include the development of microfluidic devices for quantitative analysis of proteins from single-cells, next-generation sequencing, compartmentalized organ-on-chip studies, and desalination of water on the microscale.











