When cutting a finger, thrombocytes and fibrin in the blood make up the blood-clotting mechanism, aka. haemostasis, to stop the blood loss. Another way to trigger this mechanism is having an artery damaged by atherosclerosis, which is often caused by several genetic or acquired factors. In the latter case, thrombosis develops within a vein or artery, obstructing or stopping the blood flow to major organs like the heart and eventually causing heart attack. Considering every year over 14 million lives worldwide are lost to heart attacks, more investigation on this topic is needed without any doubt.
Recently, a research team led by Andries van der Meer published a research article in Lab on a Chip on mimicking arterial thrombosis in 3D vascular structures, representing a major step forward in the development of accurate and faster methods of studying arterial thrombosis without using animals. The authors highlighted the inconvenience of using animal models to predict arterial thrombosis in humans. This is mainly due to fundamental differences between human and animal physiology, the researchers explain. For instance, rodent platelet biology, coagulation dynamics, and shear stress in mice arteries significantly vary between humans and mice.
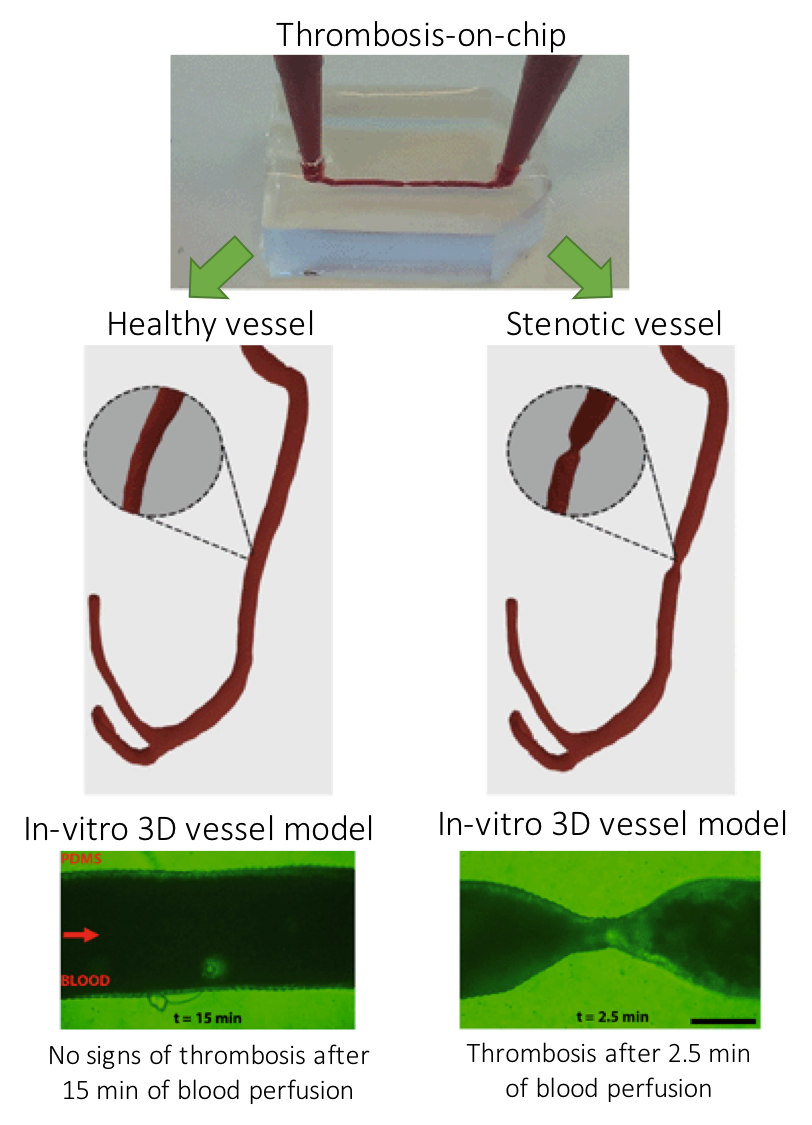
Figure 1. Three-dimensional models of a healthy and stenotic vessels and thrombosis formation upon blood perfusion through the channels.
The paper uses miniaturized vascular structures mimicking 3D architectures found in both healthy and stenotic blood vessels in-vitro (Figure 1). They combined stereolithography and 3D printing of computed tomography angiography data to construct 3D-printed templates of vessels in PDMS microchips. The 3D printed vessels are then coated with human umbilical vein endothelial cells, forming a monolayer fully covering the surface. In the next step, the artificial vessels are perfused with blood at normal arterial shear rates, allowing a blood clot to form as it would happen in the human body. The 3D printed vessel is clinically more relevant when compared to 2D vessel models, since the realistic flow profiles of blood and even distribution of shear stress across the vessel are of great importance when researching arterial thrombosis. Hugo Albers, the co-first author of the paper explains what led the team to try 3D models: “Other groups have worked on thrombosis-on-a-chip before, but we wanted to incorporate flow profiles that are similar to what one would find in-vivo. So we opt for a round and thus 3D shape. Since the stenotic geometry is an important part of this work, we wanted to find a technique that allowed us to make almost any shape we could come up with. Thus 3D-printing seemed to be the way to go.”
When it comes to defining the challenges in 3D organ-on-chip modeling and fabrication, “we needed to replicate the cellular environment using human endothelial cells and human whole blood to fully mimic the nature of vasculature” says Albers. “Incorporating the shape of vasculature to recreate the flow profiles found in-vivo and recreating the shape of vasculature on a small scale was quite challenging, since the resolution of 3D-printing quickly started to be the limiting factor. Furthermore, we ran into problems related to working with whole blood. We had to figure out how to perfuse small channels with blood without instigating thrombosis outside of the microfluidic channel.” The researchers successfully overcame the challenges mentioned by Albers and mimicked the formation of thrombosis in a stenotic vessel model as seen in Figure 1 (bottom).
The researchers note that the next step involves co-culturing arterial endothelial cells and smooth muscle cells with human umbilical vein endothelial cells or moving to different cell lines such as differentiated human induced pluripotent stem cells. “I think we can also apply the 3D-printing technique to create thrombosis-on-a-chip devices with different geometries, e.g. aneurysms or bifurcated geometries”, says Albers.
To download the full article for free* click the link below:
Pedro F. Costa, Hugo J. Albers, John E. A. Linssen, Heleen H. T. Middelkamp, Linda van der Hout, Robert Passier, Albert van den Berg, Jos Malda and Andries D. van der Meer
Lab Chip, 2017, Paper
DOI: 10.1039/C7LC00202E
This paper is included in our Organ-, Body- and Disease-on-a-Chip Thematic Collection. To read other articles in the collection, visit – rsc.li/organonachip
Burcu Gumuscu is a postdoctoral fellow in Herr Lab at UC Berkeley in the United States. Her research interests include development of microfluidic devices for quantitative analysis of proteins from single-cells, next generation sequencing, compartmentalized organ-on-chip studies, and desalination of water on the microscale.













