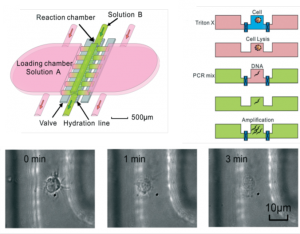
Researchers at Virginia Tech create an elegant device to perform DNA amplification starting from whole cells by taking advantage of diffusivity differences in PCR components.
Diffusion can be friend or foe in the microscale regime, depending on the application. For active mixing, relying on diffusion can lengthen reaction time and thereby decrease reaction efficiency. But for separating reaction products, low ratios of convection to diffusion (Péclet number) enable control over elements based on their diffusivity[1]. Professors Luke Achenie and Chang Lu from the chemical engineering department at Virginia Tech took advantage of this diffusion-enabled control to combine cell lysis and PCR reactions in ‘one pot’ with temporal separation of how components add to the chamber due to diffusivity differences. Separation of cell lysis and DNA amplification steps in PCR is important as many traditional chemical reagents for cell lysis inhibit polymerases used in PCR and Phusion polymerases tolerant to surfactant lysis reagents are incompatible with downstream SYBR green dyes.
The device consists of a single reaction chamber connected on both sides to two separate loading chambers. A hydration line ensures minimal evaporation during the PCR cycle in the main chamber. The loading chambers are opened in sequence to let molecules into the reaction chamber via two-layer control valves. The substantial difference in reagent diffusivity in the lysis and amplification processes allow diffusion gradients to drive molecules from new solutions contacting the reaction chamber and replace reagents from previous steps without disturbing the DNA of interest. Taq polymerase and proteins are two orders of magnitude larger in diffusivity than typical (50 kb) DNA fragments, while primers, dNTPs, and lysis buffers are three orders smaller. Relying solely on diffusion to deliver reagents to the main chamber increases the time of the reaction, but this can be addressed by elevating the temperature or increasing concentration of starting reagents in the loading chambers.
The authors showed the functionality of their device with purified human genomic DNA as well as single cells. This work opens up new capabilities to perform multi-step preparation and amplification assays for DNA in a single chamber starting directly from few cells to a single cell.
Download the full article today – for free*
Diffusion-based microfluidic PCR for “one-pot” analysis of cells
Sai Ma, Despina Nelie Loufakis, Zhenning Cao, Yiwen Chang, Luke E Achenie and Chang Lu
DOI:10.1039/C4LC00498A
References: [1] T. M. Squires and S. R. Quake, Reviews of Modern Physics, 2005, 77, 977.
*Access is free through a registered RSC account until 19th September 2014 – click here to register
About the Webwriter
Sasha is a PhD student in bioengineering working with Professor Beth Pruitt’s Microsystems lab at Stanford University. Her research focuses on evaluating relationships between cell geometry, intracellular structure, and force generation (contractility) in heart muscle cells. Outside the lab, Sasha enjoys hiking, kickboxing, and interactive science outreach.