Written by Aileen Day.
We are pleased to announce that we have just imported 1047 CIFs to ChemSpider of crystal structures that were previously reported in RSC papers (and are available as ESI for those) to ChemSpider for the relevant compounds, and linked those back to the original articles and to the CCDC’s webCSD, e.g. example compound with RSC article CIF (see the CIF infobox). Since each CIF that is uploaded into ChemSpider must be associated with a ChemSpider compound, the difficult part of this task was working out a 2D molecular structure (in .mol file format) for each 3D crystal structure (in .cif file format) – which is particularly difficult because CIFs only contain information about each atomic position and not how the atoms are bonded to each other in the crystal or whether they are charged or not.
Ultimately we would like this CIF to mol conversion (and the whole upload) to be performed programmatically without human intervention. However, there is no reliable way to do that currently – although programs such as OpenBabel can be used to extract mols from each CIF, the reliability of this conversion isn’t 100%.
So as one of our student intern projects at the University of Southampton this summer (in parallel with another student intern project at Southampton University to share thesis data in ChemSpider) we used OpenBabel (version 2.3.2, run from the command line with the options -i cif inputfilename.txt -o mol -m –unique -d –AddPolarH) to extract mols for all the CIFs in the RSC archive (over 43,000 files as of June 2013) and enlisted Julija Kezina (shown below) to review the results of these conversions to ensure that only good structure and CIF pairs would be deposited to ChemSpider, and to better understand the problems in the conversion process with a view to fixing them. One problem that became immediately apparent was that because the 2D structure obtained was just a projection of the 3D structure along the a cell axis, which is not always the orientation which shows the molecule most clearly, even if they did have the write chemical connections between the atoms, so all mol structures were run through OpenEye’s cleaning algorithm before being reviewed.
Julija compared each structure in the output mol files with those in the original CIF files to judge whether the conversion was accurate or not. In addition, as an extra check, all of the output mol structures were submitted to ChemSpider validation and standardisation platform to filter out molecules with structural problems (e.g. stereochemistry, valence or congestion issues).
Overall, approximately 30% of the CIF to mol conversions that Julija checked were good, with the right connectivity of atoms and ions (although approximately 30% of these needed the atomic positions to be repositioned to clean or tidy up the structure, either manually or using ChemDraw’s cleaning functionality). The 1047 of these mols which contain only a single molecule (without solvent molecules or cocrystals etc.) are those which have been deposited into ChemSpider with their corresponding CIFs.
The journals which had the highest successful conversion percentage were Molecular BioSystems (57%), MedChemComm (51%), Organic and Biomolecular Chemistry (44%) and Green Chemistry (44%) – the journals which in general are about small organic molecules.
Julija was working in the National Crystallography Service’s office at the University of Southampton, under the co-supervision of Professor Simon Coles, and we are grateful to them for their help and advice about the finer points of the CIF file format.
Unsuccessful CIF to mol conversions
Running and evaluating OpenBabel on such a large and varied set of structures has given us a useful opportunity to identify and categorise the most common problems encountered. Here we share these and give examples that would enable the identification of some easy fixes in the pipeline that might benefit the whole community and be used as test cases when doing so. We will report these bugs to the OpenBabel forum and because OpenBabel is open source, hope to resolve at least some of these issues in the future through collaboration with its other developers.
The following OpenBabel bugs look like they might be most straightforward to fix:
| Details | Example |
|
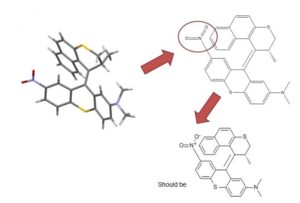
|
|
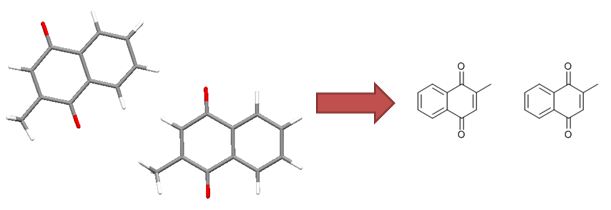
|
|

|
|

|
Many of the problems were caused by idiosynchronies or errors in the input CIFs, but these on the whole weren’t handled well by OpenBabel (e.g. by writing an error message and terminating the program) but rather, in the majority of cases went into an infinite loop and the program hung. Because of this, and because the OpenBabel conversions were part of a longer script, all OpenBabel jobs had to be run with an arbitary timeout so that if still running after this timeout they were killed, which may have discarded some valid but long-running OpenBabel jobs. We will investigate whether there is a validation program that can be automatically performed on CIFs to filter out ones with these problems (similar to the CCDC’s EnCIFer but which can be run programmatically), but it would be relatively straightforward to make OpenBabel more reliable by being able to exit nicely when it encounters these problems so that pre-validation wasn’t necessary. These problems are listed in the table below:
| Details | Example |
|
|
|

|
|

|
The following OpenBabel bugs were the most frequent in occurence, but will be difficult to fix. They arise from the problem that the CIF format does not record charges on atoms/ions or the types of bong between them so OpenBabel needs to work them out which is hard to do correctly.
| Details | Example |
|
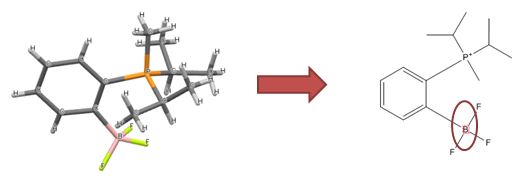
|
|
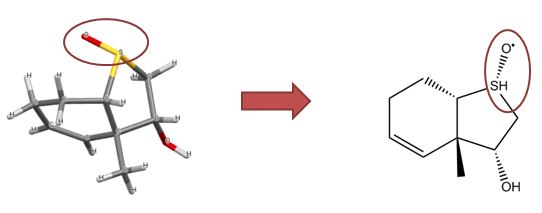
|
|
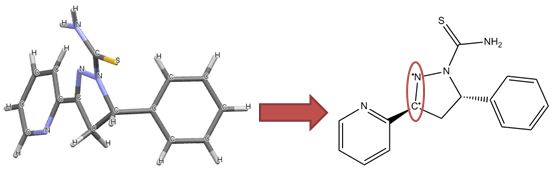
|
|
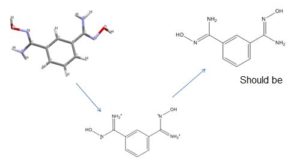
|
|
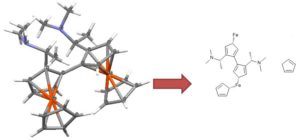
|
|
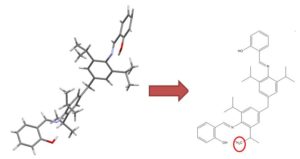
|
There were also some problem mol files produced which either won’t be able to be fixed by OpenBabel (since they resulted from either errors or limitations of the input CIF files which cannot be fixed retrospectively) or are too difficult to fix and/or too infrequently occuring to be worth the effort:
-
- There were 237 cases where there were solvent molecules in the CIF (many of which have missing hydrogens, partial occupancy of the molecule or part of the molecule etc.) which give rise to spurious oxygens, fragments of molecules and radicals in the resulting mol file (see CIF: CCDC 213787 and ChemSpider record: 68005706). 148 of these cases are just water solvent molecules either with missing or detached hydrogen atoms. The poor definition of the solvent molecules is a limitation of CIF files from diffraction so it is not possible for OpenBabel to better define them in the output mol that is derived from them. However, running OpenBabel with the -r option to remove all but the largest contiguous fragment was quite successful to remove these problem solvent molecules so no further action is required to deal with this problem and this option will be used by us in the future.
- There were 81 cases where there was at least one missing hydrogen in the original CIF (or in 3 cases, all hydrogens missing) – see CCDC 259871.
- Some CIFs contain crystal structures which correspond to continuous networks rather than small molecules (e.g. polymers, MOFs, zeolites, POMs) which cannot meaningfully be captured in mol format – see CCDC 206593.
- There were a few (24) cases where the stereochemistry in the mol file obtained is incorrectly defined. However, because on the stereochemistry was well interpreted by OpenBabel and these cases were relatively few, it probably isn’t worth disturbing the apple cart to investigate these further – see CCDC 238611 and ChemSpider 9419187.


