The affordable and rapid testing of DNA hybridization and polymorphisms is crucial for the diagnosis of genetic and infectious diseases. DNA biosensors based on nucleic recognition represent one of the most recent technologies currently employed. Electrochemical transducers are commonly used for sensing of DNA hybridization because they are inexpensive, small and highly sensitive. However, this kind of device sees the attachment of a single DNA probe onto the electrode surface to act as recognition element for the complementary target, and an external electro-active label that recognises this complex structure is required. To avoid the use of an external electro-active label, researchers have started to use new materials.
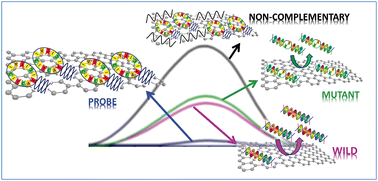
Differential pulse voltammetry to examine the oxidation of guanine on target DNA
Adeline Huiling Loo and co-workers, from the Nanyang Technological University of Singapore, used graphene, a low cost substance made of a single layer of carbon atoms densely packed. Graphene has high electron conductivity, large specific surface area and fast electron transfer, and is considered a material with a great potential for biosening applications.
The authors analysed the properties of graphene oxide, electrochemically reduced graphene oxide and thermally reduced graphene oxide and investigated for the first time the mechanisms behind physically immobilized hairpin DNA hybridization. For the label free detection of DNA hybridization and polymorphism, they employed differential pulse voltammetry and examined the oxidation of guanine on target DNA molecules hybridized with an inosine-substituted hairpin DNA probe. According to the study, graphene oxides show the best performance in terms of recognition of complementary and non-complementary DNA sequences.
To know more about this research, click on the link below. This paper will be free to read until January 9th.
An insight into the hybridization mechanism of hairpin DNA physically immobilized on chemically modified graphenes
Adeline Huiling Loo , Alessandra Bonanni and Martin Pumera
Analyst, 2013, 138, 467-471
DOI: 10.1039/C2AN36199J