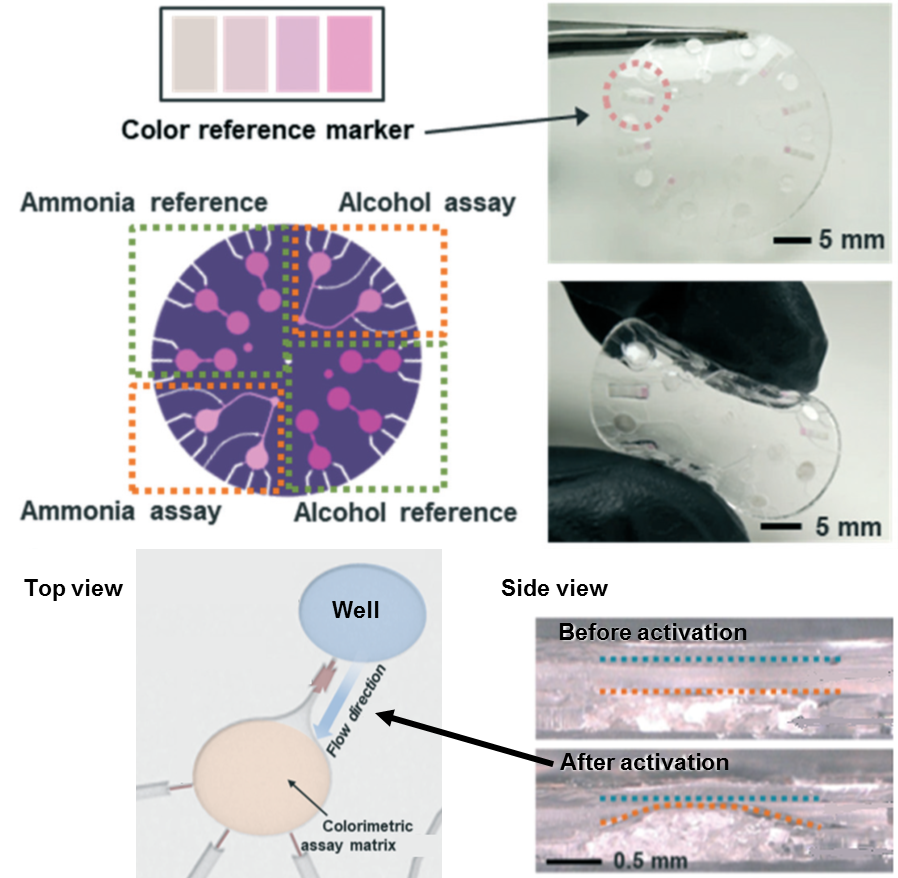
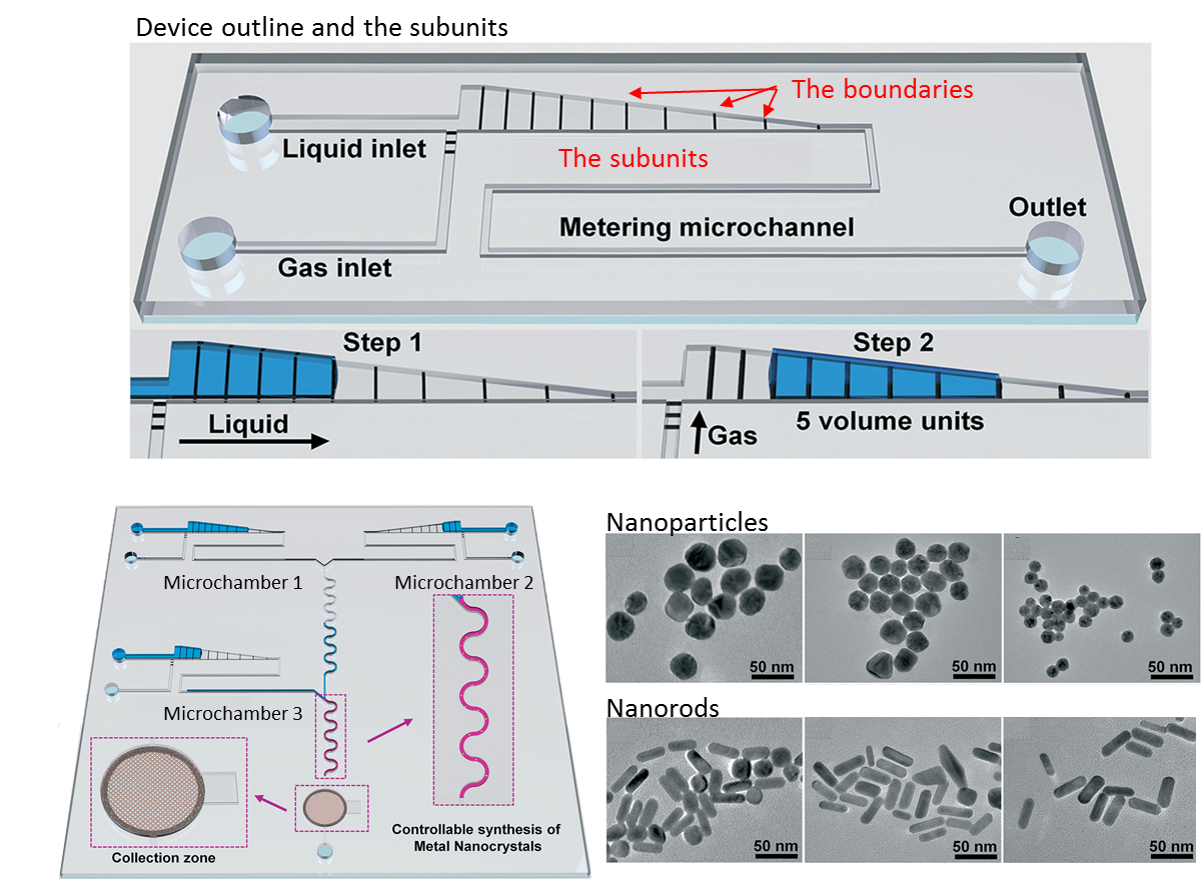
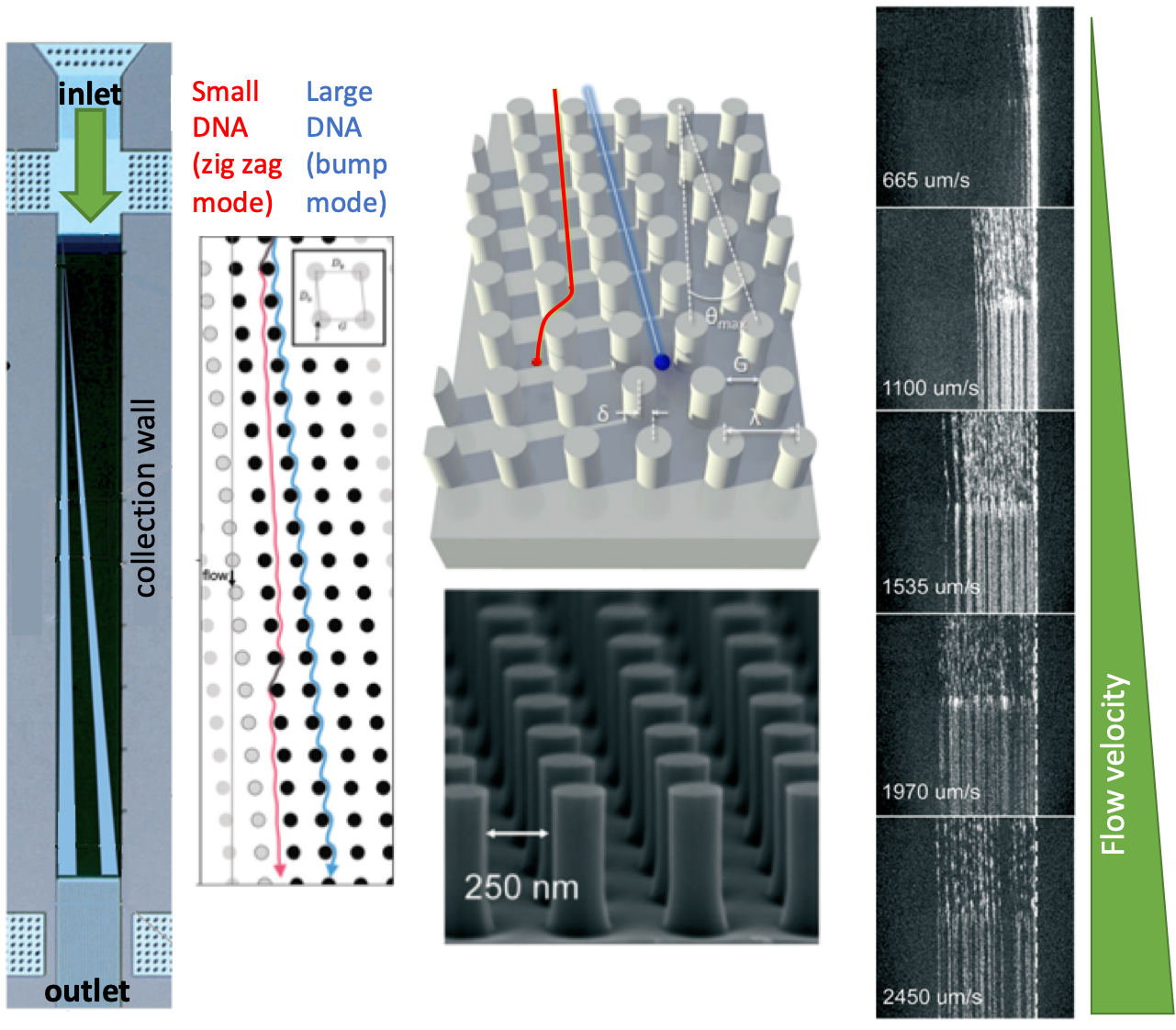
Creating a lab-on-chip device with multiple sensing capabilities has been a long-desired goal in the biotechnology field. A sensor-rich chip would pack the power of a full-scale laboratory as initially envisioned in the lab on a chip concept, yet it remained challenging for decades. Many researchers suggest that real-time monitoring of the culture conditions can provide higher-quality drug screening testing. Although many platforms have been proposed up till now, none of them has yet taken off. A recent work from Andreas Weltin’s laboratory at IMTEK published in the Lab on a Chip, however, sounds very promising.
Embedding the cells in synthetic or natural 3D matrices makes it possible to mimic an in vivo cellular environment, but also frequently takes away control over the culturing conditions. The present microfluidic platform achieves the spheroid growth of tumor cells using Matrigel for several days. However, without continuous monitoring, large amounts of independent cultures would be necessary, as each must be sacrificed to conduct separate assays at separate time points, raising the degree of uncertainty in statistical analysis. The authors addressed this problem by integrating several sensors which can perform continuous readout of different molecules.
Creating hypoxia (low oxygen levels) or hyperoxia (high oxygen levels) conditions in cell culture is often needed to mimic disease conditions for fundamental studies. On the other hand, monitoring the local oxygen levels in a culture is not straightforward. In the present study, the authors fabricated the reference electrodes by electroplating silver/silver chloride. The oxygen sensors were modified using a platinum-coated surface located at multiple spots in close vicinity of the culture chamber, allowing for a comparison of the initial and waste streams (Figure 1).
Cell metabolism underpins cell activity, as well as its viability. Some of the key parameters to monitor cell metabolism include glucose and lactate concentrations in the culture media. While high uptake of glucose is indicative of faster cell respiration and replication, high production of lactate is indicative of an increased anaerobic respiration rate. The authors embedded enzymatic sensors in hydrogel to measure glucose and lactate levels. Regions with lactate oxidase and glucose oxidase enzymes are strategically located in close vicinity to the culture chamber. Such locations were ideal because the cell metabolites released into the media are not yet diffused to a lower concentration at this spot, while it is far enough from the chamber so the by-products of the enzymatic reaction cannot interfere with the cell culture (Figure 1).
As a proof-of-concept, Weltin’s team applied this multifunctional sensing platform to the growth of patient-derived triple-negative breast cancer stem cells, which are highly metastatic and less responsive to treatment. The developed platform unveiled the difference in temporal and concentration-dependent drug response of the cells in spheroids. The authors state that this work underlines the importance of in situ, real-time metabolite monitoring in 3D cell cultures as a future standard in cancer research-related studies.
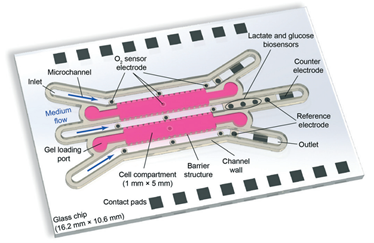
Figure 1. The layout of the microfluidic chip. The authors created a compartmentalized matrix-embedded cell culture with medium perfusion and rigorous control of the liquid and gas composition in one platform but also incorporated continuous sensors that are capable to deliver real-time readouts of the oxygen concentrations and cell metabolism by-products.
About the Web writers
 Burcu Gumuscu is an assistant professor at Eindhoven University of Technology in the Netherlands, and the chair of the Biosensors and Devices Laboratory. She strives for the development, fabrication, and application of smart biomaterials to realize high-precision processing in high-throughput microfluidic settings. She specifically focuses on the design and development of lab-on-a-chip devices containing hydrogels for diversified life sciences applications.
Burcu Gumuscu is an assistant professor at Eindhoven University of Technology in the Netherlands, and the chair of the Biosensors and Devices Laboratory. She strives for the development, fabrication, and application of smart biomaterials to realize high-precision processing in high-throughput microfluidic settings. She specifically focuses on the design and development of lab-on-a-chip devices containing hydrogels for diversified life sciences applications.
Oksana Savchak is a Ph.D. student in Biosensors and Devices Laboratory at the Eindhoven University of Technology in the Netherlands. She focuses on the development of microfluidic screening platforms to investigate cell-material interactions.
Original publication
Dornhof, J., Kieninger, J., Muralidharan, H., Maurer, J., Urban, G. A., Weltin, A. (2022): Microfluidic organ-on-chip system for multi-analyte monitoring of metabolites in 3D cell cultures. In: Lab on a Chip, 22 (2). DOI: 10.1039/d1lc00689d