Ayako Yamada123, Jean-Louis Viovy123, Catherine Villard123 and Stéphanie Descroix123
1 Laboratoire Physico Chimie Curie, Institut Curie, PSL Research University, CNRS UMR168, Paris, France.
2 Sorbonne Universités, UMPC Univ. Paris 06, Paris, France
3 Institut Pierre-Gilles de Gennes, Paris, France
Email: ayako.yamada@curie.fr
Why is this useful?
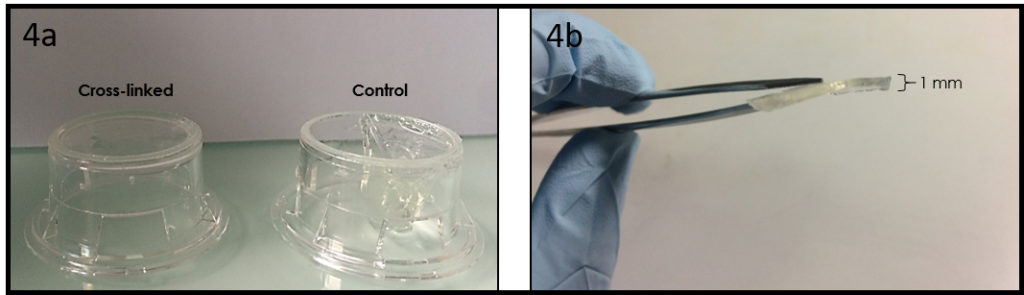
Glass is a versatile surface for chemical treatments, and it still is by far the most used substrate for surface engineering (e.g. micropatterning, surface chemistry) or plasma-bonding of PDMS microfluidic devices. For cell culture on such substrates, glass-bottom culture dishes are desired to keep over the cells well defined medium volumes, and to protect the cells from contamination and medium evaporation. Moreover, they are optically better suited for microscopy observation than polystyrene dishes routinely used for cell culture. Although glass-bottom culture dishes are commercially available (e.g. Fluorodish from WPI), the presence of plastic walls limits the treatments that can be performed onto the glass bottom surface and those are more expensive ( 5 € per dish; φ 50 mm) than polystyrene dishes (e.g. φ 40 mm dish from TPP, 0.5 € per dish). In this Tip, we describe an easier way than a previous Tip1 to transform a polystyrene culture dish into a glass-bottom one, while preserving the possibility to apply to the glass any treatment before its assembly into a dish. Note that in this method, the body of the culture dish will be upside down and the lid is thus no longer lifted above the dish opening by lid stoppers. However, the gas exchange through the gap between the body and its lid seems to be enough to culture cells healthily in this dish. In summary, this Tip provides a low-cost and rapid solution for cell culture in a microfluidic device or on an engineered surface directly in a culture dish, suited for a long-term live cell imaging.
What do I need?
- φ 40 mm polystyrene tissue culture dish (e.g. TPP #93040)
- φ 40 mm cover slide (e.g. Thermo Fisher Scientific #11757065, ∼0.2 € per slide)
- Large screw driver (or a similar tool)
- Uncured mixture of PDMS base and curing agent (10:1 w/w)
- Oven or hotplate
- Ferromagnetic metal plate (e.g. lid of a PDMS container, optional)
- Cylindrical magnets (optional)
What do I do?
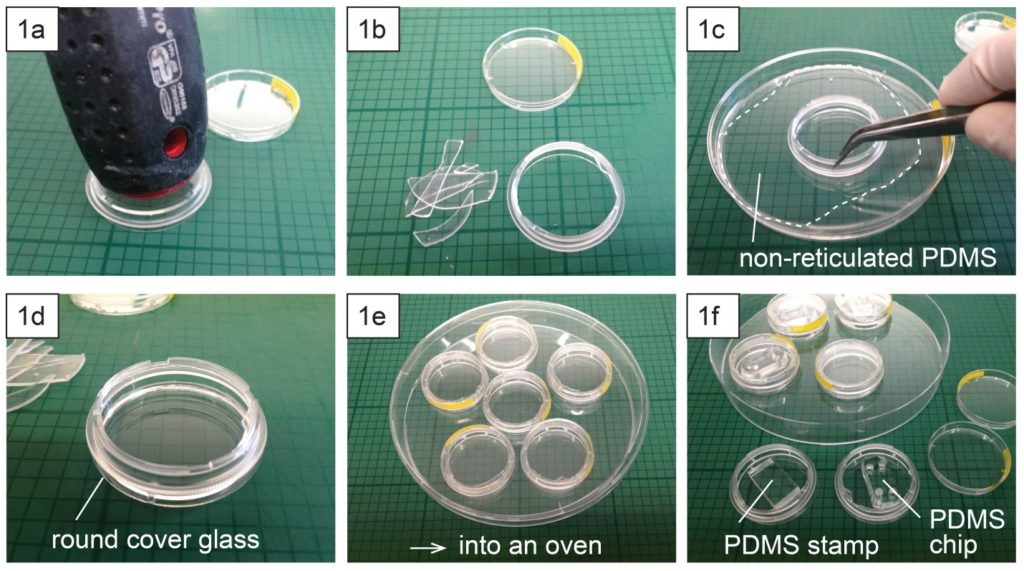
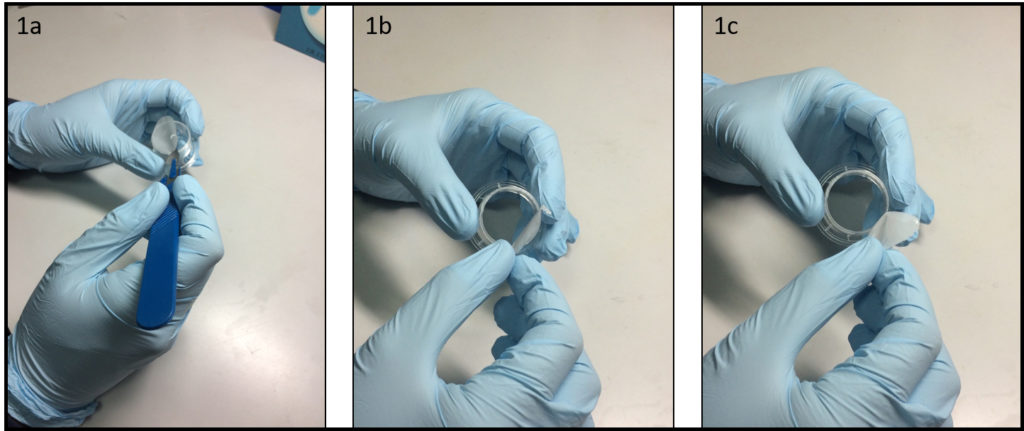
- Place a polystyrene culture dish upside down on a surface and hit a few times the center of the dish bottom with the grip of a large screw driver (Fig. 1a) until the dish bottom falls apart from the dish wall (Fig. 1b). The bottom should fall easily with the success rate around 9 over 10. Avoid breaking the dish wall by hitting the bottom too strongly.
- Spread uncured PDMS mixture on a flat substrate (e.g. a larger plastic Petri dish) and coat the edge of the dish (broken part up) with PDMS (Fig. 1c).
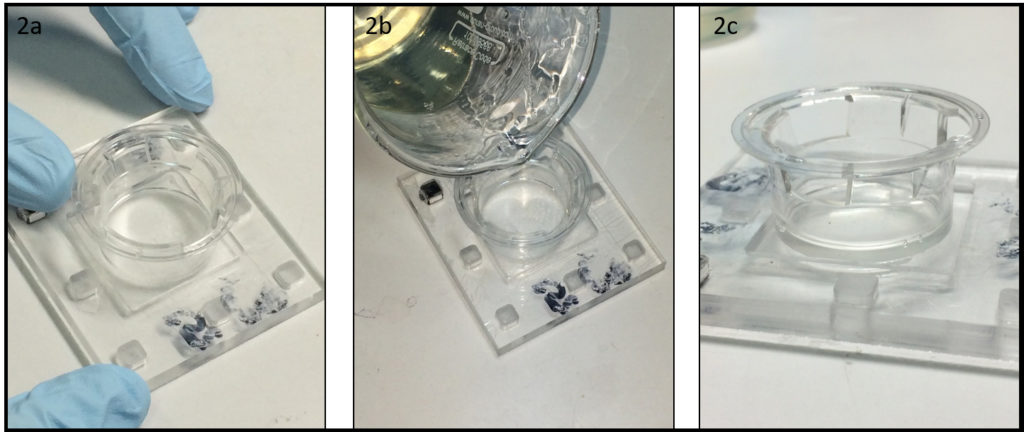
- Place the dish (broken part up) on a cover glass slide (Fig. 1d) and cure PDMS in an oven or on a hotplate e.g. at 80 °C for 10 min (Fig. 1e).
- Surface treatment (e.g. micro-contact printing) or plasma-bonding of a PDMS chip to the glass surface can be performed after or before the dish assembly (Fig. 1f).
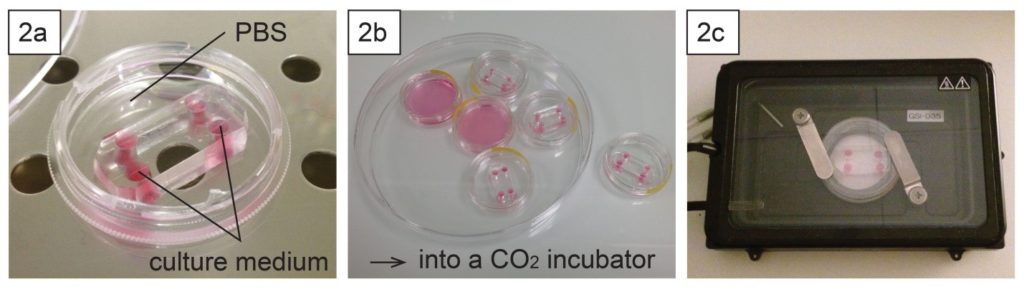
- To keep humidity for on-chip cell culture, the dish can be filled with e.g. phosphate buffered saline (Fig. 2a). Dishes with chips or micropatterns loaded with cells can be placed in a CO2 incubator with or without further protection (Fig. 2b).
- Long-term live cell imaging can be performed using a stage top incubator (Fig. 2c).
What else should I know?
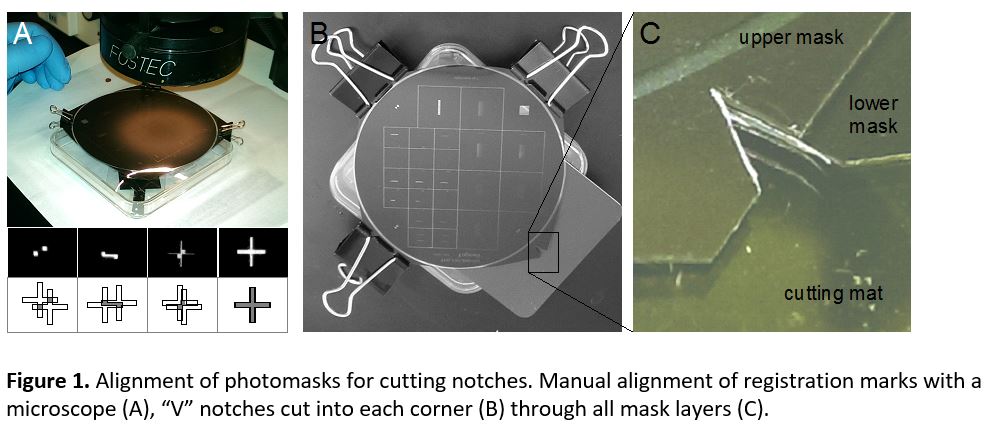
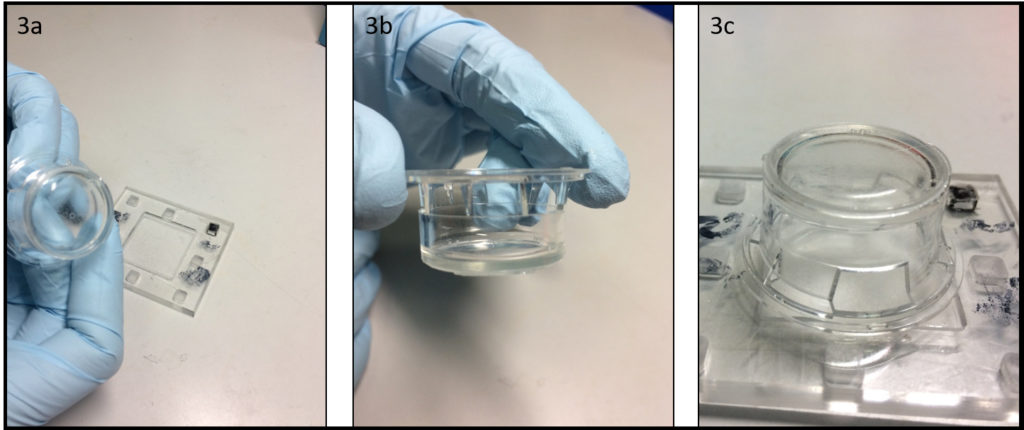
- Depending on the support type of microscopes, it might be necessary to well align the contours of the dish and the glass slide. This can be done using cylindrical magnets (3 per dish) and a ferromagnetic metal plate (Fig. 3a) during PDMS curing in an oven or on a hotplate (Fig. 3b).
Acknowledgement
This work is supported by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” program (reference: ANR 10-NANO 0207) and ERC Advanced Grant CellO (FP7-IDEAS-ERC-321107).
Reference
[1] Caballero D, Samitier J, Different strategies for the fabrication of cell culture chambers for live-cell imaging studies. Chips and Tips, 02 Dec 2014 (https://blogs.rsc.org/chipsandtips/2014/12/02/different-strategies-for-the-fabrication-of-cell-culture-chambers-for-live-cell-imaging-studies/)