We are delighted to share the latest selections in the Molecular Omics Editor’s collection. This is a showcase of some of the best articles published in our journal, hand selected by our Associate Editors and Editorial Board members.
 This selection is from our Associate Editor Richard Unwin. Richard graduated from the University of Nottingham with a BSc in Biology and MSc in Oncology before obtaining his PhD from the University of Leeds in 2001 in what was then the new field of Proteomics. He subsequently joined the University of Manchester, developing new methods for analysing cancer proteomes, including isobaric tagging, global and targeted analysis of protein phosphorylation, and methods for comparing proteomic and transcriptomic data.
This selection is from our Associate Editor Richard Unwin. Richard graduated from the University of Nottingham with a BSc in Biology and MSc in Oncology before obtaining his PhD from the University of Leeds in 2001 in what was then the new field of Proteomics. He subsequently joined the University of Manchester, developing new methods for analysing cancer proteomes, including isobaric tagging, global and targeted analysis of protein phosphorylation, and methods for comparing proteomic and transcriptomic data.
In 2010 he moved to manage a new mass spectrometry research laboratory within the UK National Health Service, where he worked on the study of proteins and metabolites in chronic disease before returning to the University of Manchester in 2017 to continue a research programme developing tools for mass spectrometry data acquisition and analysis for the study of age-related chronic diseases.
Professor Unwin has highlighted some of their favourite recent articles below:
 Integrative analysis of cancer dependency data and comprehensive phosphoproteomics data revealed the EPHA2-PARD3 axis as a cancer vulnerability in KRAS-mutant colorectal cancer
Integrative analysis of cancer dependency data and comprehensive phosphoproteomics data revealed the EPHA2-PARD3 axis as a cancer vulnerability in KRAS-mutant colorectal cancer
Daigo Gunji, Ryohei Narumi, Satoshi Muraoka, Junko Isoyama, Narumi Ikemoto, Mimiko Ishida, Takeshi Tomonaga, Yoshiharu Sakai, Kazutaka Obama and Jun Adachi
Mol. Omics, 2023, 19, 624-639
Richard’s comments
“Colorectal cancer (CRC) remains one of the most untreatable cancers. Here, the authors provide an in-depth analysis of the proteome and phosphoproteome of 35 CRC cell lines in order to correlate protein expression or signalling with mutation status in the KRAS and BRAF genes. Their study identified specific protein interactions and signalling pathways that are associated with specific genotypes, and further identified a series of tight junction-related proteins associated with hard-to-treat KRAS mutant cancers.”
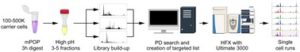 Single cell proteomics analysis of drug response shows its potential as a drug discovery platform
Single cell proteomics analysis of drug response shows its potential as a drug discovery platform
Juerg Straubhaar, Alexandria D’Souza, Zachary Niziolek and Bodgan Budnik
Mol. Omics, 2024, 20, 6-18
Richard’s comments
“Single cell-based omics, in particular proteomics, are enhancing our fundamental understand of biology but provide unique challenges around speed and sensitivity. As such, methods are evolving rapidly as the field moves forward. This paper adds to this effort by modifying a well-established workflow to increase penetration into the single-cell proteome and provides good use cases for the technology in model systems.”
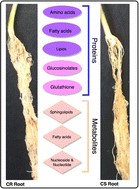 Proteome- and metabolome-level changes during early stages of clubroot infection in Brassica napus canola
Proteome- and metabolome-level changes during early stages of clubroot infection in Brassica napus canola
Dinesh Adhikary, Devang Mehta, Anna Kisiala, Urmila Basu, R. Glen Uhrig, R. J. Neil Emery, Habibur Rahman and Nat N. V. Kav
Mol. Omics, 2024, 20, 265-282
Richard’s comments
“Canola is an important oilseed crop, yet worldwide yields are impacted significantly by infection with clubroot. In this paper, the authors perform an integrated proteomic and metabolomics analysis of infection-resistant and susceptible strains and provide the first look at the molecular changes in the root as a result of infection. Data integration and pathway analysis reveals a number of molecules which appear to be important in protection of infection, and these provide targets for future strains created either via gene editing or as markers for future strain development.”
Enjoyed these articles? Check out our latest publications and if you want the chance to be part of the next edition of our Editor’s collection, submit your research here.











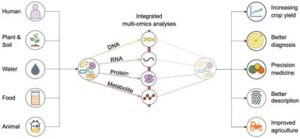 Integrated multi-omics analyses of microbial communities: a review of the current state and future directions
Integrated multi-omics analyses of microbial communities: a review of the current state and future directions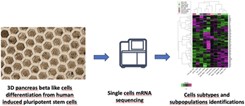 Generation of β-like cell subtypes from differentiated human induced pluripotent stem cells in 3D spheroids
Generation of β-like cell subtypes from differentiated human induced pluripotent stem cells in 3D spheroids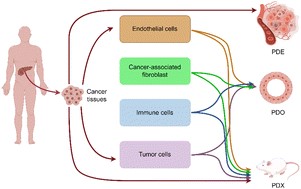 Pancreatic cancer environment: from patient-derived models to single-cell omics
Pancreatic cancer environment: from patient-derived models to single-cell omics