Written by Colin Batchelor.
You might not think so, but you’re very good at taking a two-dimensional drawing and converting it into a three-dimensional shape in your head. No, really, you are.
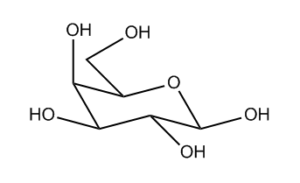
Take the drawing of galatose in Fig. 1. Even if you’re not a chemist, you can tell which bits of the ring are at the front and at the back, which bonds point up and which bonds point down. If you actually are a chemist, you’ve been trained to apply this geometrical intuition to work out what’s going on at each of the five stereocentres.
However, if you ask the InChI algorithm about the stereochemistry of this molecule, it’ll say that there is no stereochemistry in there and you’re looking at a stereoless description of which atom is attached to which. Since we use the InChI algorithm to say whether two records describe the same molecule, this puts us in a quandary, and there are thousands of entries in ChemSpider that come from just such a drawing and hence lack stereochemistry.
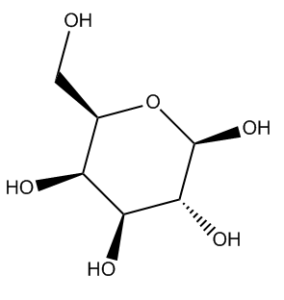
I’ve been developing algorithms to take these allegedly flat drawings and render them in such a way that InChI and other algorithms can identify the stereochemistry and turn the side-on view above into the eagle’s-eye view in Fig. 2.
Currently we can standardize chair sugars and Haworth-projection sugars, and this is being built into CVSP. More on our methods and what we will be doing with them, not least tidying up the erroneous ChemSpider entries, soon!