Recent advances in our understanding of RNA have expanded its role beyond just a carrier of genetic information. Modern views of RNA encompass a diverse range of molecules, which play a central role in modulating a number of important biological processes, including the progression of various cancers and nontumorigenic diseases.
In her recent OBC publication, Professor Amanda Hargrove of Duke University has created a general, high throughput screening technique for the identification of small molecule inhibitors of different RNA proteins using a fluorescence indicator displacement (FID) assay.
Small molecule RNA inhibitors offer advantages such as enhanced absorption, distribution, and oral bioavailability over commonly employed oligonucleotides used in antisense technologies. Fluorescence-based assays are widely used techniques used to screen small molecule libraries, however, the incorporation of fluorescent tags can lead to alterations in the RNA’s native structure and often pose synthetic challenges. FID, on the other hand, is a ‘tagless’ approach in which the indicator displays different fluorescent properties in the presence or absence of an oligonucleotide.
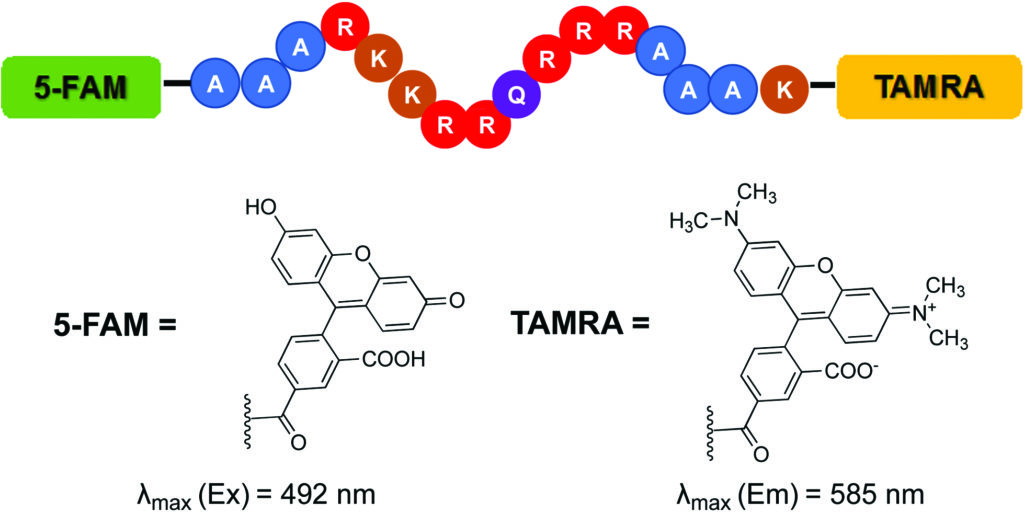
The Hargrove group implemented the Tat FID assay in order to simultaneously screen a library of small molecules against multiple RNA targets. This provides an efficient means of measuring binding affinity as well as evaluating specificity. The Tat FID assay requires a Tat peptide construct labeled with a Förster Resonance Enhancement Transfer (FRET) pair—a 5-carboxyfluorescein (FAM) at the N-terminus and 5-carboxytetramethylrhodamine (TAMRA) at the C-terminus (see figure). When the peptide is bound to RNA, the distance between fluorophores allows for excitation of FAM and emission detection from TAMRA. Displacement of the Tat peptide results in quenching of the fluorescent signal, which can be used to quantify binding affinity.
A library of 30 small molecules (including known RNA binding small molecules) was screened against four select RNA structures. The assay was successful in identifying hits for all four RNA molecules and identified indiscriminate and differential binding of individual small molecules. Binding results correlated with known literature results and statistical analysis was used to clarify the relationship between small molecule structures/RNA based on their binding affinity and selectivity.
As with any fluorescence-based assay, the Tat FID assay possesses inherent limitations. However, it has been shown to be a powerful high throughput technique as a result of its ease of operation, low concentration requirement for both ligand and target, and its ability to rapidly assess small molecule/RNA binding interactions.
To find out more see:
Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA
Neeraj N. Patwardhan,
DOI:10.1039/C8OB02467G
Victoria Corless has recently completed her Ph.D. in organic chemistry with Prof. Andrei Yudin at the University of Toronto. Her research is centered on the synthesis of kinetically amphoteric building blocks which offer a versatile platform for the development of chemoselective transformations with particular emphasis on creating novel biologically active molecules. She is passionate about communicating new discoveries to enhance science literacy.