Cell behavior is highly dependent on its surrounding environment, including neighboring and adjacent cells. Individual cells can merge and form a solid-like state for a “jamming” effect or a highly dense cell mass can disperse and become more mobile, for an “unjamming” transition. Both behaviors have been observed in complex, multi-cellular interactions such as wound healing, embryonic development, and tumor metastasis.
To investigate jamming-unjamming cell transitions, researchers from Brown University describe methods to quantify cell-cell interactions in a recently published Soft Matter article. The group observed co-cultures of epithelial cells and mesenchymal cells and their ability to cluster (jamming) or remain mobile and unconnected (unjamming). Epithelial cells collectively organize to form dense multi-layered cell sheets while mesenchymal cells avoid cell-cell bonds and are capable of individual migration.
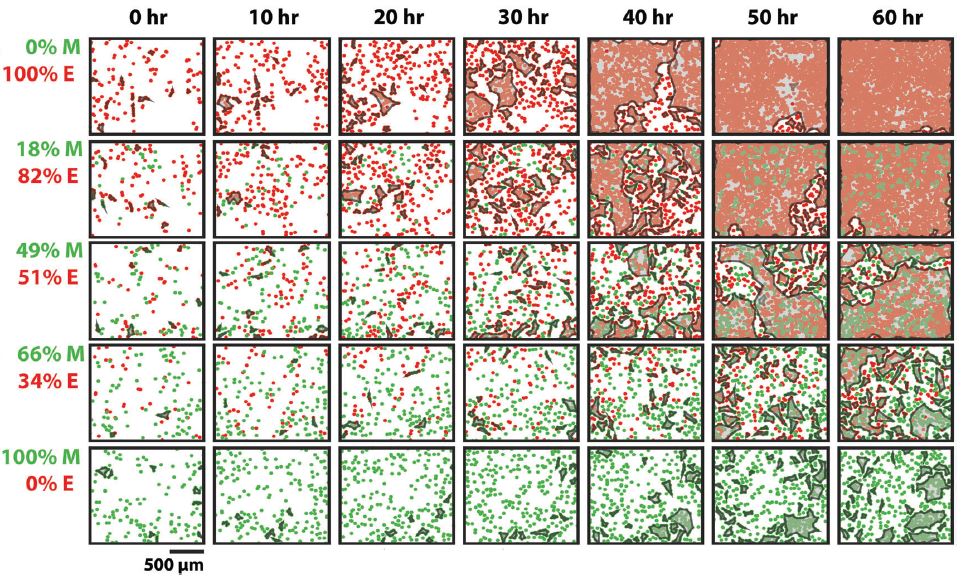
Cell clustering ability with different ratios of epithelial (red) and mesenchymal cells (green)
By increasing the number of mesenchymal cells in an epithelial cell population, the research group observed a reduction in epithelial cell clustering causing a significant disruption in cell sheet formation and confluency. The addition of mesenchymal cells also increased the average collective cell velocity and decreased cell proliferation, reducing the typical jamming behavior of epithelial cells.
The research provides important biophysical data for collective cell behavior as well as introducing new parameters to control cell jamming-unjamming transitions.
Interested in this research? Read the full article for free until 31/10/2016 using a registered RSC account:
Clustering and jamming in epithelial-mesenchymal co-cultures
Marielena Gamboa Castro, Susan E. Leggett, and Ian Y. Wong
Soft Matter, 2016, Advance Article
DOI: 10.1039/C6SM01287F
—————-
Dr. Morgan M. Stanton is currently a postdoctoral researcher at the Max Planck Institute for Intelligent Systems in Stuttgart, Germany. Her research focuses on the cell-material interface material and properties regulating cell behavior.
Read more about Morgan’s research publications and follow her on Twitter: @morg368.